Reciprocal modulation of mesenchymal stem cells and tumor cells promotes lung cancer metastasis
- PMID: 29503225
- PMCID: PMC5925622
- DOI: 10.1016/j.ebiom.2018.02.017
Reciprocal modulation of mesenchymal stem cells and tumor cells promotes lung cancer metastasis
Abstract
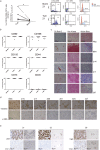
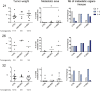
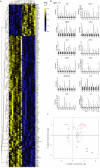
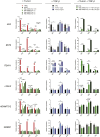
Metastasis is a multi-step process in which direct crosstalk between cancer cells and their microenvironment plays a key role. Here, we assessed the effect of paired tumor-associated and normal lung tissue mesenchymal stem cells (MSCs) on the growth and dissemination of primary human lung carcinoma cells isolated from the same patients. We show that the tumor microenvironment modulates MSC gene expression and identify a four-gene MSC signature that is functionally implicated in promoting metastasis. We also demonstrate that tumor-associated MSCs induce the expression of genes associated with an aggressive phenotype in primary lung cancer cells and selectively promote their dissemination rather than local growth. Our observations provide insight into mechanisms by which the stroma promotes lung cancer metastasis.
Keywords: ADAMTS12; GREM1; ITGA11; LOXL2; Tumor-associated MSCs; lung cancer; metastasis.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Oncostatin m modulates the mesenchymal-epithelial transition of lung adenocarcinoma cells by a mesenchymal stem cell-mediated paracrine effect.Cancer Res. 2012 Nov 15;72(22):6051-64. doi: 10.1158/0008-5472.CAN-12-1568. Epub 2012 Nov 8. Cancer Res. 2012. PMID: 23139208
-
Roles of cell fusion between mesenchymal stromal/stem cells and malignant cells in tumor growth and metastasis.FEBS J. 2021 Mar;288(5):1447-1456. doi: 10.1111/febs.15483. Epub 2020 Jul 24. FEBS J. 2021. PMID: 33070450 Review.
-
CXCR1/Akt signaling activation induced by mesenchymal stem cell-derived IL-8 promotes osteosarcoma cell anoikis resistance and pulmonary metastasis.Cell Death Dis. 2018 Jun 18;9(7):714. doi: 10.1038/s41419-018-0745-0. Cell Death Dis. 2018. PMID: 29915309 Free PMC article.
-
HIF-1α promoted vasculogenic mimicry formation in hepatocellular carcinoma through LOXL2 up-regulation in hypoxic tumor microenvironment.J Exp Clin Cancer Res. 2017 Apr 27;36(1):60. doi: 10.1186/s13046-017-0533-1. J Exp Clin Cancer Res. 2017. PMID: 28449718 Free PMC article.
-
The multifaceted role of mesenchymal stem cells in cancer.Semin Cancer Biol. 2020 Feb;60:225-237. doi: 10.1016/j.semcancer.2019.06.003. Epub 2019 Jun 15. Semin Cancer Biol. 2020. PMID: 31212021 Review.
Cited by
-
Clinical Roles of Risk Model Based on Differentially Expressed Genes in Mesenchymal Stem Cells in Prognosis and Immunity of Non-small Cell Lung Cancer.Front Genet. 2022 Feb 24;13:823075. doi: 10.3389/fgene.2022.823075. eCollection 2022. Front Genet. 2022. PMID: 35281822 Free PMC article.
-
The therapeutic potential of stem cell-derived exosomes in the ulcerative colitis and colorectal cancer.Stem Cell Res Ther. 2022 Apr 1;13(1):138. doi: 10.1186/s13287-022-02811-5. Stem Cell Res Ther. 2022. PMID: 35365226 Free PMC article. Review.
-
Prognostic Epstein-Barr Virus (EBV) miRNA biomarkers for survival outcome in EBV-associated epithelial malignancies: Systematic review and meta-analysis.PLoS One. 2022 Apr 18;17(4):e0266893. doi: 10.1371/journal.pone.0266893. eCollection 2022. PLoS One. 2022. PMID: 35436288 Free PMC article.
-
Stem Cell-Induced Cell Motility: A Removable Obstacle on the Way to Safe Therapies?Stem Cells Transl Med. 2022 Mar 3;11(1):26-34. doi: 10.1093/stcltm/szab003. Stem Cells Transl Med. 2022. PMID: 35641164 Free PMC article.
-
Mesenchymal stem cells in cancer progression and anticancer therapeutic resistance.Cancer Cell Int. 2021 Nov 4;21(1):595. doi: 10.1186/s12935-021-02300-4. Cancer Cell Int. 2021. PMID: 34736460 Free PMC article. Review.
References
-
- Aggarwal S., Pittenger M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815–1822. - PubMed
-
- Alamgeer M., Ganju V., Watkins D.N. Novel therapeutic targets in non-small cell lung cancer. Curr. Opin. Pharmacol. 2013;13:394–401. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

