Bacterial microcompartments
- PMID: 29503457
- PMCID: PMC6022854
- DOI: 10.1038/nrmicro.2018.10
Bacterial microcompartments
Abstract
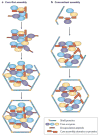
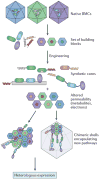
Bacterial microcompartments (BMCs) are self-assembling organelles that consist of an enzymatic core that is encapsulated by a selectively permeable protein shell. The potential to form BMCs is widespread and found across the kingdom Bacteria. BMCs have crucial roles in carbon dioxide fixation in autotrophs and the catabolism of organic substrates in heterotrophs. They contribute to the metabolic versatility of bacteria, providing a competitive advantage in specific environmental niches. Although BMCs were first visualized more than 60 years ago, it is mainly in the past decade that progress has been made in understanding their metabolic diversity and the structural basis of their assembly and function. This progress has not only heightened our understanding of their role in microbial metabolism but is also beginning to enable their use in a variety of applications in synthetic biology. In this Review, we focus on recent insights into the structure, assembly, diversity and function of BMCs.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures



References
-
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. - PubMed
-
- Kerfeld CA, Melnicki MR. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol. 2016;31:66–75. - PubMed
-
- Turmo A, Gonzalez-Esquer CR, Kerfeld CA. Carboxysomes: metabolic modules for CO2 fixation. FEMS Microbiol Lett. 2017;364 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

