Comparative Genomic Analysis of a Clinical Isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 Beta-Lactamases Producer Harboring Two Drug-Resistance Plasmids from Southeast Brazil
- PMID: 29503635
- PMCID: PMC5820359
- DOI: 10.3389/fmicb.2018.00220
Comparative Genomic Analysis of a Clinical Isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 Beta-Lactamases Producer Harboring Two Drug-Resistance Plasmids from Southeast Brazil
Abstract
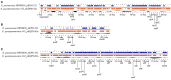
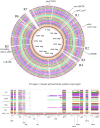
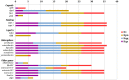
The aim of this study was to unravel the genetic determinants responsible for multidrug (including carbapenems) resistance and virulence in a clinical isolate of Klebsiella quasipneumoniae subsp. similipneumoniae by whole-genome sequencing and comparative analyses. Eighty-three clinical isolates initially identified as carbapenem-resistant K. pneumoniae were collected from nosocomial infections in southeast Brazil. After RAPD screening, the KPC-142 isolate, showing the most divergent DNA pattern, was selected for complete genome sequencing in an Illumina HiSeq 2500 instrument. Reads were assembled into scaffolds, gaps between scaffolds were resolved by in silico gap filling and extensive bioinformatics analyses were performed, using multiple comparative analysis tools and databases. Genome sequencing allowed to correct the classification of the KPC-142 isolate as K. quasipneumoniae subsp. similipneumoniae. To the best of our knowledge this is the first complete genome reported to date of a clinical isolate of this subspecies harboring both class A beta-lactamases KPC-2 and OKP-B-6 from South America. KPC-142 has one 5.2 Mbp chromosome (57.8% G+C) and two plasmids: 190 Kbp pKQPS142a (50.7% G+C) and 11 Kbp pKQPS142b (57.3% G+C). The 3 Kbp region in pKQPS142b containing the blaKPC-2 was found highly similar to that of pKp13d of K. pneumoniae Kp13 isolated in Southern Brazil in 2009, suggesting the horizontal transfer of this resistance gene between different species of Klebsiella. KPC-142 additionally harbors an integrative conjugative element ICEPm1 that could be involved in the mobilization of pKQPS142b and determinants of resistance to other classes of antimicrobials, including aminoglycoside and silver. We present the completely assembled genome sequence of a clinical isolate of K. quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 beta-lactamases producer and discuss the most relevant genomic features of this important resistant pathogen in comparison to several strains belonging to K. quasipneumoniae subsp. similipneumoniae (phylogroup II-B), K. quasipneumoniae subsp. quasipneumoniae (phylogroup II-A), K. pneumoniae (phylogroup I), and K. variicola (phylogroup III). Our study contributes to the description of the characteristics of a novel K. quasipneumoniae subsp. similipneumoniae strain circulating in South America that currently represent a serious potential risk for nosocomial settings.
Keywords: KPC-2; Klebsiella quasipneumoniae subsp similipneumoniae; OKP-B-6; complete genome sequence; nosocomial infection; silver resistance.
Figures


Similar articles
-
First Report of Whole-Genome Sequence of Colistin-Resistant Klebsiella quasipneumoniae subsp. similipneumoniae Producing KPC-9 in India.Microb Drug Resist. 2019 May;25(4):489-493. doi: 10.1089/mdr.2018.0116. Epub 2018 Nov 14. Microb Drug Resist. 2019. PMID: 30427763
-
Characterisation of clinical carbapenem-resistant K1 Klebsiella quasipneumoniae subsp. similipneumoniae strains harbouring a virulence plasmid.Int J Antimicrob Agents. 2022 Aug;60(2):106628. doi: 10.1016/j.ijantimicag.2022.106628. Epub 2022 Jun 24. Int J Antimicrob Agents. 2022. PMID: 35760224
-
Whole genome sequencing reveals antibiotic resistance pattern and virulence factors in Klebsiella quasipneumoniae subsp. Similipneumoniae from Hospital wastewater in South-West, Nigeria.Microb Pathog. 2024 Dec;197:107040. doi: 10.1016/j.micpath.2024.107040. Epub 2024 Oct 19. Microb Pathog. 2024. PMID: 39427715
-
Characterization of a multidrug-resistant hypovirulent ST1859-KL35 klebsiella quasipneumoniae subsp. similipneumoniae strain co-harboring tmexCD2-toprJ2 and blaKPC-2.J Glob Antimicrob Resist. 2025 May;42:253-261. doi: 10.1016/j.jgar.2025.03.009. Epub 2025 Mar 18. J Glob Antimicrob Resist. 2025. PMID: 40113085
-
Genomic and phenotypic characterization of ST2012 clinical Klebsiella quasipneumoniae subsp. similipneumoniae harboring blaNDM-1 in China.BMC Microbiol. 2024 Nov 29;24(1):506. doi: 10.1186/s12866-024-03637-2. BMC Microbiol. 2024. PMID: 39614148 Free PMC article.
Cited by
-
Complete Genomic Analysis of a Kingdom-Crossing Klebsiella variicola Isolate.Front Microbiol. 2018 Oct 9;9:2428. doi: 10.3389/fmicb.2018.02428. eCollection 2018. Front Microbiol. 2018. PMID: 30356723 Free PMC article.
-
Molecular Characterization of Carbapenem Resistant Klebsiella pneumoniae and Klebsiella quasipneumoniae Isolated from Lebanon.Sci Rep. 2019 Jan 24;9(1):531. doi: 10.1038/s41598-018-36554-2. Sci Rep. 2019. PMID: 30679463 Free PMC article.
-
Genomic insights of beta-lactamase producing Klebsiella quasipneumoniae subsp. similipneumoniae belonging to sequence type 1699 from retail market fish, India.Arch Microbiol. 2022 Jul 4;204(8):454. doi: 10.1007/s00203-022-03071-w. Arch Microbiol. 2022. PMID: 35781844
-
Complete Genome Sequence of Klebsiella quasipneumoniae subsp. similipneumoniae Strain IF3SW-P1, Isolated from the International Space Station.Microbiol Resour Announc. 2022 Jul 21;11(7):e0047622. doi: 10.1128/mra.00476-22. Epub 2022 Jun 23. Microbiol Resour Announc. 2022. PMID: 35735981 Free PMC article.
-
Genomics of Klebsiella pneumoniae Species Complex Reveals the Circulation of High-Risk Multidrug-Resistant Pandemic Clones in Human, Animal, and Environmental Sources.Microorganisms. 2022 Nov 17;10(11):2281. doi: 10.3390/microorganisms10112281. Microorganisms. 2022. PMID: 36422351 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

