Prevalence and Genetic Basis of Antimicrobial Resistance in Non- aureus Staphylococci Isolated from Canadian Dairy Herds
- PMID: 29503642
- PMCID: PMC5820348
- DOI: 10.3389/fmicb.2018.00256
Prevalence and Genetic Basis of Antimicrobial Resistance in Non- aureus Staphylococci Isolated from Canadian Dairy Herds
Abstract
Emergence and spread of antimicrobial resistance is a major concern for the dairy industry worldwide. Objectives were to determine: (1) phenotypic and genotypic prevalence of drug-specific resistance for 25 species of non-aureus staphylococci, and (2) associations between presence of resistance determinants and antimicrobial resistance. Broth micro-dilution was used to determine resistance profiles for 1,702 isolates from 89 dairy herds. Additionally, 405 isolates were sequenced to screen for resistance determinants. Antimicrobial resistance was clearly species-dependent. Resistance to quinupristin/dalfopristin was common in Staphylococcus gallinarum (prevalence of 98%), whereas S. cohnii and S. arlettae were frequently resistant to erythromycin (prevalence of 63 and 100%, respectively). Prevalence of resistance was 10% against β-lactams and tetracyclines. In contrast, resistance to antimicrobials critically important for human medicine, namely vancomycin, fluoroquinolones, linezolid and daptomycin, was uncommon (< 1%). Genes encoding multidrug-resistance efflux pumps and resistance-associated residues in deducted amino acid sequences of the folP gene were the most frequent mechanisms of resistance, regardless of species. The estimated prevalence of the mecA gene was 17% for S. epidermidis. Several genes, including blaZ, mecA, fexA, erm, mphC, msrA, and tet were associated with drug-specific resistance, whereas other elements were not. There were specific residues in gyrB for all isolates of species intrinsically resistant to novobiocin. This study provided consensus protein sequences of key elements previously associated with resistance for 25 species of non-aureus staphylococci from dairy cattle. These results will be important for evaluating effects of interventions in antimicrobial use in Canadian dairy herds.
Keywords: antimicrobial resistance; antimicrobial resistance genes; coagulase-negative staphylococci; dairy; mastitis; non-aureus staphylococci; prevalence.
Figures
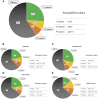

References
-
- Aarestrup F. M., Seyfarth A. M., Emborg H. D., Pedersen K., Hendriksen R. S., Bager F. (2001). Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45, 2054–2059. 10.1128/AAC.45.7.2054-2059.2001 - DOI - PMC - PubMed
-
- Antiabong J. F., Kock M. M., Bellea N. M., Ehlers M. M. (2017). Diversity of multidrug efflux genes and phenotypic evaluation of the in vitro resistance dynamics of clinical Staphylococcus aureus isolates using methicillin; a model beta-lactam. Open Microbiol. J. 11, 132–141. 10.2174/1874285801711010132 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

