Gene expression in retinal ischemic post-conditioning
- PMID: 29504043
- PMCID: PMC5911419
- DOI: 10.1007/s00417-018-3905-0
Gene expression in retinal ischemic post-conditioning
Abstract
Purpose: The pathophysiology of retinal ischemia involves mechanisms including inflammation and apoptosis. Ischemic post-conditioning (Post-C), a brief non-lethal ischemia, induces a long-term ischemic tolerance, but the mechanisms of ischemic post-conditioning in the retina have only been described on a limited basis. Accordingly, we conducted this study to determine the molecular events in retinal ischemic post-conditioning and to identify targets for therapeutic strategies for retinal ischemia.
Methods: To determine global molecular events in ischemic post-conditioning, a comprehensive study of the transcriptome of whole retina was performed. We utilized RNA sequencing (RNA-Seq), a recently developed, deep sequencing technique enabling quantitative gene expression, with low background noise, dynamic detection range, and discovery of novel genes. Rat retina was subjected to ischemia in vivo by elevation of intraocular pressure above systolic blood pressure. At 24 h after ischemia, Post-C or sham Post-C was performed by another, briefer period of ischemia, and 24 h later, retinas were collected and RNA processed.
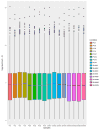
Results: There were 71 significantly affected pathways in post-conditioned/ischemic vs. normals and 43 in sham post conditioned/ischemic vs. normals. Of these, 28 were unique to Post-C and ischemia. Seven biological pathways relevant to ischemic injury, in Post-C as opposed to sham Post-C, were examined in detail. Apoptosis, p53, cell cycle, JAK-STAT, HIF-1, MAPK and PI3K-Akt pathways significantly differed in the number as well as degree of fold change in genes between conditions.
Conclusion: Post-C is a complex molecular signaling process with a multitude of altered molecular pathways. We identified potential gene candidates in Post-C. Studying the impact of altering expression of these factors may yield insight into new methods for treating or preventing damage from retinal ischemic disorders.
Keywords: Ischemia; Post-conditioning; RNA-Seq; Retina.
Figures






References
-
- Zheng L, Gong B, Hatala DA, Kern TS. Retinal ischemia and reperfusion causes capillary degeneration: similarities to diabetes. Invest Ophthalmol Vis Sci. 2007;48:361–367. - PubMed
-
- Fernandez DC, Bordone MP, Chianelli MS, Rosenstein RE. Retinal neuroprotection against ischemia-reperfusion damage induced by postconditioning. Invest Ophthalmol Vis Sci. 2009;50:3922–3930. - PubMed
-
- Osborne NN, Ugarte M, Chao M, Chidlow G, Bae JH, Wood JP, Nash MS. Neuroprotection in relation to retinal ischemia and relevance to glaucoma. Surv Ophthalmol. 1999;43(Suppl 1):S102–128. - PubMed
-
- Limalanathan S, Andersen GO, Hoffmann P, Klow N-E, Abdelnoor M, Eritsland J. Rationale and design of the POSTEMI (postconditioning in ST-elevation myocardial infarction) study. Cardiology. 2010;116:103–109. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

