Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants
- PMID: 29504210
- PMCID: PMC6097130
- DOI: 10.1111/pbi.12896
Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants
Abstract
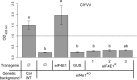
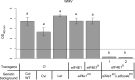
To infect plants, viruses rely heavily on their host's machinery. Plant genetic resistances based on host factor modifications can be found among existing natural variability and are widely used for some but not all crops. While biotechnology can supply for the lack of natural resistance alleles, new strategies need to be developed to increase resistance spectra and durability without impairing plant development. Here, we assess how the targeted allele modification of the Arabidopsis thaliana translation initiation factor eIF4E1 can lead to broad and efficient resistance to the major group of potyviruses. A synthetic Arabidopsis thaliana eIF4E1 allele was designed by introducing multiple amino acid changes associated with resistance to potyvirus in naturally occurring Pisum sativum alleles. This new allele encodes a functional protein while maintaining plant resistance to a potyvirus isolate that usually hijacks eIF4E1. Due to its biological functionality, this synthetic allele allows, at no developmental cost, the pyramiding of resistances to potyviruses that selectively use the two major translation initiation factors, eIF4E1 or its isoform eIFiso4E. Moreover, this combination extends the resistance spectrum to potyvirus isolates for which no efficient resistance has so far been found, including resistance-breaking isolates and an unrelated virus belonging to the Luteoviridae family. This study is a proof-of-concept for the efficiency of gene engineering combined with knowledge of natural variation to generate trans-species virus resistance at no developmental cost to the plant. This has implications for breeding of crops with broad-spectrum and high durability resistance using recent genome editing techniques.
Keywords: Arabidopsis thaliana; eIF4E; potyvirus; resistance; synthetic allele; translational research.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures







References
-
- Andrade, M. , Abe, Y. , Nakahara, K.S. and Uyeda, I. (2009) The cyv‐2 resistance to Clover yellow vein virus in pea is controlled by the eukaryotic initiation factor 4E. J. Gen. Plant Pathol. 75, 241–249.
-
- Baltes, N.J. and Voytas, D.F. (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33, 120–131. - PubMed
-
- Barabaschi, D. , Tondelli, A. , Desiderio, F. , Volante, A. , Vaccino, P. , Valè, G. and Cattivelli, L. (2016) Next generation breeding. Plant Sci. 242, 3–13. - PubMed
-
- Bastet, A. , Robaglia, C. and Gallois, J.L. (2017) eIF4E resistance: Natural variation should guide gene editing. Trends Plant Sci. 22, 411–419. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

