Comparing miRNA structure of mirtrons and non-mirtrons
- PMID: 29504892
- PMCID: PMC5836839
- DOI: 10.1186/s12864-018-4473-8
Comparing miRNA structure of mirtrons and non-mirtrons
Abstract
Background: MicroRNAs proceeds through the different canonical and non-canonical pathways; the most frequent of the non-canonical ones is the splicing-dependent biogenesis of mirtrons. We compare the mirtrons and non-mirtrons of human and mouse to explore how their maturation appears in the precursor structure around the miRNA.
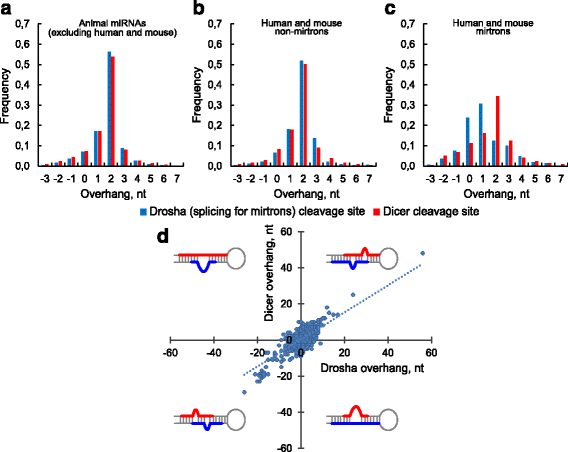
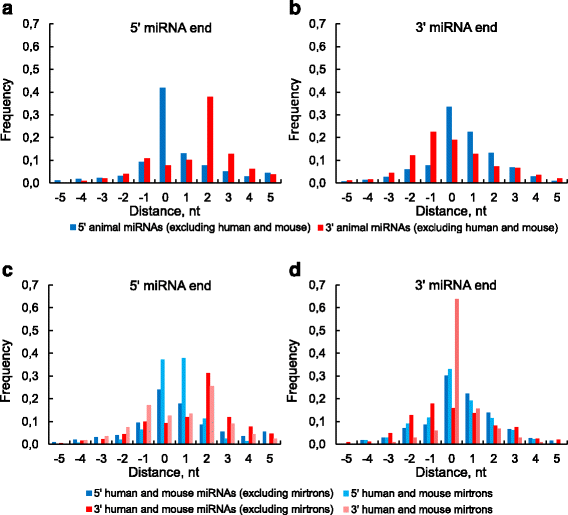
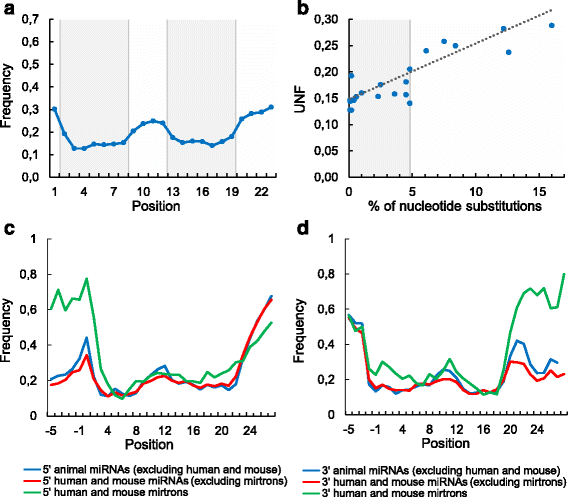
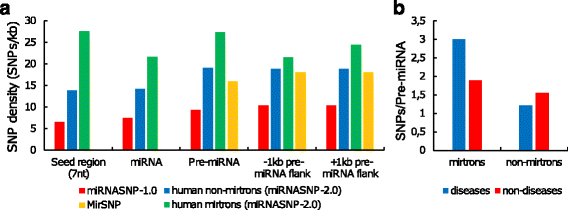
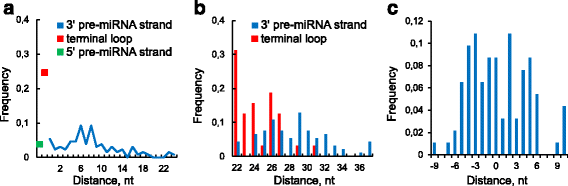
Results: We found the coherence of the overhang lengths what indicates the dependence between the cleavage sites. To explain this dependence we suggest the 2-lever model of the Dicer structure that couples the imprecisions in Drosha and Dicer. Considering the secondary structure of all animal pre-miRNAs we confirmed that single-stranded nucleotides tend to be located near the miRNA boundaries and in its center and are characterized by a higher mutation rate. The 5' end of the canonical 5' miRNA approaches the nearest single-stranded nucleotides what suggests the extension of the loop-counting rule from the Dicer to the Drosha cleavage site. A typical structure of the annotated mirtron pre-miRNAs differs from the canonical pre-miRNA structure and possesses the 1- and 2 nt hanging ends at the hairpin base. Together with the excessive variability of the mirtron Dicer cleavage site (that could be partially explained by guanine at its ends inherited from splicing) this is one more evidence for the 2-lever model. In contrast with the canonical miRNAs the mirtrons have higher snp densities and their pre-miRNAs are inversely associated with diseases. Therefore we supported the view that mirtrons are under positive selection while canonical miRNAs are under negative one and we suggested that mirtrons are an intrinsic source of silencing variability which produces the disease-promoting variants. Finally, we considered the interference of the pre-miRNA structure and the U2snRNA:pre-mRNA basepairing. We analyzed the location of the branchpoints and found that mirtron structure tends to expose the branchpoint site what suggests that the mirtrons can readily evolve from occasional hairpins in the immediate neighbourhood of the 3' splice site.
Conclusion: The miRNA biogenesis manifests itself in the footprints of the secondary structure. Close inspection of these structural properties can help to uncover new pathways of miRNA biogenesis and to refine the known miRNA data, in particular, new non-canonical miRNAs may be predicted or the known miRNAs can be re-classified.
Keywords: Biogenesis; Branchpoint; Dicer structure; Mirtron; Overhangs; Secondary structure; Splicing; miRNA; microRNA.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

