Features of the organization of bread wheat chromosome 5BS based on physical mapping
- PMID: 29504906
- PMCID: PMC5836826
- DOI: 10.1186/s12864-018-4470-y
Features of the organization of bread wheat chromosome 5BS based on physical mapping
Abstract
Background: The IWGSC strategy for construction of the reference sequence of the bread wheat genome is based on first obtaining physical maps of the individual chromosomes. Our aim is to develop and use the physical map for analysis of the organization of the short arm of wheat chromosome 5B (5BS) which bears a number of agronomically important genes, including genes conferring resistance to fungal diseases.
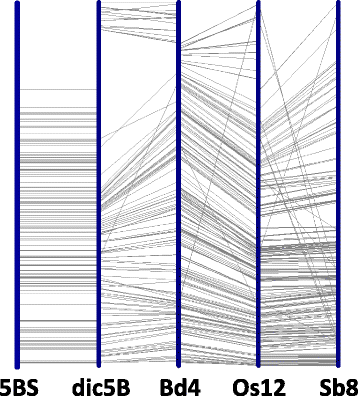
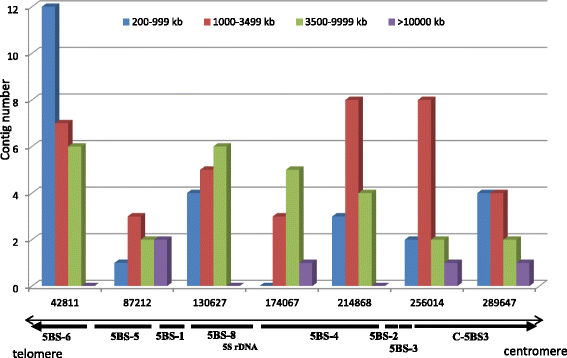
Results: A physical map of the 5BS arm (290 Mbp) was constructed using restriction fingerprinting and LTC software for contig assembly of 43,776 BAC clones. The resulting physical map covered ~ 99% of the 5BS chromosome arm (111 scaffolds, N50 = 3.078 Mb). SSR, ISBP and zipper markers were employed for anchoring the BAC clones, and from these 722 novel markers were developed based on previously obtained data from partial sequencing of 5BS. The markers were mapped using a set of Chinese Spring (CS) deletion lines, and F2 and RICL populations from a cross of CS and CS-5B dicoccoides. Three approaches have been used for anchoring BAC contigs on the 5BS chromosome, including clone-by-clone screening of BACs, GenomeZipper analysis, and comparison of BAC-fingerprints with in silico fingerprinting of 5B pseudomolecules of T. dicoccoides. These approaches allowed us to reach a high level of BAC contig anchoring: 96% of 5BS BAC contigs were located on 5BS. An interesting pattern was revealed in the distribution of contigs along the chromosome. Short contigs (200-999 kb) containing markers for the regions interrupted by tandem repeats, were mainly localized to the 5BS subtelomeric block; whereas the distribution of larger 1000-3500 kb contigs along the chromosome better correlated with the distribution of the regions syntenic to rice, Brachypodium, and sorghum, as detected by the Zipper approach.
Conclusion: The high fingerprinting quality, LTC software and large number of BAC clones selected by the informative markers in screening of the 43,776 clones allowed us to significantly increase the BAC scaffold length when compared with the published physical maps for other wheat chromosomes. The genetic and bioinformatics resources developed in this study provide new possibilities for exploring chromosome organization and for breeding applications.
Keywords: Chromosome 5BS; Genetic markers; Hexaploid wheat; Physical mapping; Sequencing; Synteny; Triticum aestivum.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Whole-genome profiling and shotgun sequencing delivers an anchored, gene-decorated, physical map assembly of bread wheat chromosome 6A.Plant J. 2014 Jul;79(2):334-47. doi: 10.1111/tpj.12550. Epub 2014 Jun 19. Plant J. 2014. PMID: 24813060 Free PMC article.
-
Physical mapping of a large plant genome using global high-information-content-fingerprinting: the distal region of the wheat ancestor Aegilops tauschii chromosome 3DS.BMC Genomics. 2010 Jun 17;11:382. doi: 10.1186/1471-2164-11-382. BMC Genomics. 2010. PMID: 20553621 Free PMC article.
-
Fine organization of genomic regions tagged to the 5S rDNA locus of the bread wheat 5B chromosome.BMC Plant Biol. 2017 Nov 14;17(Suppl 1):183. doi: 10.1186/s12870-017-1120-5. BMC Plant Biol. 2017. PMID: 29143604 Free PMC article.
-
Integrated physical map of bread wheat chromosome arm 7DS to facilitate gene cloning and comparative studies.N Biotechnol. 2019 Jan 25;48:12-19. doi: 10.1016/j.nbt.2018.03.003. Epub 2018 Mar 8. N Biotechnol. 2019. PMID: 29526810
-
Genetic, physical, and comparative map of the subtelomeric region of mouse Chromosome 4.Mamm Genome. 2002 Jan;13(1):5-19. doi: 10.1007/s0033501-2109-8. Mamm Genome. 2002. PMID: 11773963 Free PMC article. Review.
Cited by
-
Physical location of tandem repeats in the wheat genome and application for chromosome identification.Planta. 2019 Mar;249(3):663-675. doi: 10.1007/s00425-018-3033-4. Epub 2018 Oct 24. Planta. 2019. PMID: 30357506
-
Novel genomic regions on chromosome 5B controlling wheat powdery mildew seedling resistance under Egyptian conditions.Front Plant Sci. 2023 May 10;14:1160657. doi: 10.3389/fpls.2023.1160657. eCollection 2023. Front Plant Sci. 2023. PMID: 37235018 Free PMC article.
-
Genomics at Belyaev conference - 2017.BMC Genomics. 2018 Feb 9;19(Suppl 3):79. doi: 10.1186/s12864-018-4476-5. BMC Genomics. 2018. PMID: 29504918 Free PMC article. No abstract available.
-
Nuclear Disposition of Alien Chromosome Introgressions into Wheat and Rye Using 3D-FISH.Int J Mol Sci. 2019 Aug 25;20(17):4143. doi: 10.3390/ijms20174143. Int J Mol Sci. 2019. PMID: 31450653 Free PMC article.
-
Dissection of Structural Reorganization of Wheat 5B Chromosome Associated With Interspecies Recombination Suppression.Front Plant Sci. 2022 May 4;13:884632. doi: 10.3389/fpls.2022.884632. eCollection 2022. Front Plant Sci. 2022. PMID: 36340334 Free PMC article.
References
-
- Clouse JW, Adhikary D, Page JT, Ramaraj T, Deyholos MK, Udall JA, et al. The Amaranth genome: genome, transcriptome, and physical map assembly. Plant Genome. 2016;9(1):1-14. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

