Reading the tea leaves: Dead transposon copies reveal novel host and transposon biology
- PMID: 29505560
- PMCID: PMC5860798
- DOI: 10.1371/journal.pbio.2005470
Reading the tea leaves: Dead transposon copies reveal novel host and transposon biology
Abstract
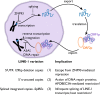
Transposable elements comprise a huge portion of most animal genomes. Unlike many pathogens, these elements leave a mark of their impact via their insertion into host genomes. With proper teasing, these sequences can relay information about the evolutionary history of transposons and their hosts. In a new publication, Larson and colleagues describe a previously unappreciated density of long interspersed element-1 (LINE-1) sequences that have been spliced (LINE-1 and other reverse transcribing elements are necessarily intronless). They provide data to suggest that the retention of these potentially deleterious splice sites in LINE-1 results from the sites' overlap with an important transcription factor binding site. These spliced LINE-1s (i.e., spliced integrated retrotransposed elements [SpiREs]) lose their ability to replicate, suggesting they are evolutionary dead ends. However, the lethality of this splicing could be an efficient means of blocking continued replication of LINE-1. In this way, the record of inactive LINE-1 sequences in the human genome revealed a new, though infrequent, event in the LINE-1 replication cycle and motivates future studies to test whether splicing might be another weapon in the anti-LINE-1 arsenal of host genomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Comment on
-
Spliced integrated retrotransposed element (SpIRE) formation in the human genome.PLoS Biol. 2018 Mar 5;16(3):e2003067. doi: 10.1371/journal.pbio.2003067. eCollection 2018 Mar. PLoS Biol. 2018. PMID: 29505568 Free PMC article.
References
-
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284(5757):601–3. . - PubMed
-
- Orgel LE, Crick FH, Sapienza C. Selfish DNA. Nature. 1980;288(5792):645–6. . - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062 . - DOI - PubMed
-
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384 doi: 10.1371/journal.pgen.1002384 . - DOI - PMC - PubMed
-
- Smit A, Hubley, R & Green, P. RepeatMasker Open-4.0 2013-2015: http://www.repeatmasker.org.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

