A novel ex vivo Huntington's disease model for studying GABAergic neurons and cell grafts by laser microdissection
- PMID: 29505597
- PMCID: PMC5837106
- DOI: 10.1371/journal.pone.0193409
A novel ex vivo Huntington's disease model for studying GABAergic neurons and cell grafts by laser microdissection
Abstract
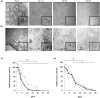
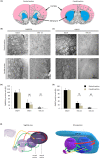
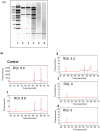
Organotypic brain slice cultures have been recently used to study neurodegenerative disorders such as Parkinson's disease and Huntington's disease (HD). They preserve brain three-dimensional architecture, synaptic connectivity and brain cells microenvironment. Here, we developed an innovative model of Huntington's disease from coronal rat brain slices, that include all the areas involved in the pathology. HD-like neurodegeneration was obtained in only one week, in a single step, during organotypic slice preparation, without the use of neurotoxins. HD-like histopathology was analysed and after one week, a reduction of 40% of medium spiny neurons was observed. To analyse new therapeutic approaches in this innovative HD model, we developed a novel protocol of laser microdissection to isolate and analyse by RT-qPCR, grafted cells as well as surrounding tissue of fresh organotypic slices. We determined that laser microdissection could be performed on a 400μm organotypic slice after alcohol dehydration protocol, allowing the analysis of mRNA expression in the rat tissue as well as in grafted cells. In conclusion, we developed a new approach for modeling Huntington's disease ex vivo, and provided a useful innovative method for screening new potential therapies for neurodegenerative diseases especially when associated with laser microdissection.
Conflict of interest statement
Figures




Similar articles
-
Profilin-2 increased expression and its altered interaction with β-actin in the striatum of 3-nitropropionic acid-induced Huntington's disease in rats.Neuroscience. 2014 Dec 5;281:216-28. doi: 10.1016/j.neuroscience.2014.09.035. Epub 2014 Sep 27. Neuroscience. 2014. PMID: 25255934
-
Neuronal properties, in vivo effects, and pathology of a Huntington's disease patient-derived induced pluripotent stem cells.Stem Cells. 2012 Sep;30(9):2054-62. doi: 10.1002/stem.1135. Stem Cells. 2012. PMID: 22628015
-
Transplantation of GABAergic cells derived from bioreactor-expanded human neural precursor cells restores motor and cognitive behavioral deficits in a rodent model of Huntington's disease.Cell Transplant. 2013;22(12):2237-56. doi: 10.3727/096368912X658809. Epub 2012 Nov 1. Cell Transplant. 2013. PMID: 23127784
-
Stem cell-based therapy for Huntington's disease.J Cell Biochem. 2013 Apr;114(4):754-63. doi: 10.1002/jcb.24432. J Cell Biochem. 2013. PMID: 23097329 Review.
-
Organotypic cultures as tools for optimizing central nervous system cell therapies.Exp Neurol. 2013 Oct;248:429-40. doi: 10.1016/j.expneurol.2013.07.012. Epub 2013 Jul 27. Exp Neurol. 2013. PMID: 23899655 Review.
Cited by
-
A Combinatorial Cell and Drug Delivery Strategy for Huntington's Disease Using Pharmacologically Active Microcarriers and RNAi Neuronally-Committed Mesenchymal Stromal Cells.Pharmaceutics. 2019 Oct 12;11(10):526. doi: 10.3390/pharmaceutics11100526. Pharmaceutics. 2019. PMID: 31614758 Free PMC article.
-
The Use of ex Vivo Rodent Platforms in Neuroscience Translational Research With Attention to the 3Rs Philosophy.Front Vet Sci. 2018 Jul 19;5:164. doi: 10.3389/fvets.2018.00164. eCollection 2018. Front Vet Sci. 2018. PMID: 30073174 Free PMC article. Review.
References
-
- Squitieri F, Griguoli A, Capelli G, Porcellini A, D’Alessio B. Epidemiology of Huntington disease: first post- HTT gene analysis of prevalence in Italy: Prevalence of Huntington disease in Italy. Clin Genet. 2015; n/a-n/a. doi: 10.1111/cge.12574 - DOI - PubMed
-
- Tepper JM, Koós T, Wilson CJ. GABAergic microcircuits in the neostriatum. Trends Neurosci. 2004;27: 662–669. doi: 10.1016/j.tins.2004.08.007 - DOI - PubMed
-
- Indersmitten T, Tran CH, Cepeda C, Levine MS. Altered excitatory and inhibitory inputs to striatal medium-sized spiny neurons and cortical pyramidal neurons in the Q175 mouse model of Huntington’s disease. J Neurophysiol. 2015;113: 2953–2966. doi: 10.1152/jn.01056.2014 - DOI - PMC - PubMed
-
- Shannon KM, Fraint A. Therapeutic advances in Huntington’s Disease. Mov Disord Off J Mov Disord Soc. 2015; doi: 10.1002/mds.26331 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

