Phosphoethanolamine-N-methyltransferase is a potential biomarker for the diagnosis of P. knowlesi and P. falciparum malaria
- PMID: 29505599
- PMCID: PMC5837800
- DOI: 10.1371/journal.pone.0193833
Phosphoethanolamine-N-methyltransferase is a potential biomarker for the diagnosis of P. knowlesi and P. falciparum malaria
Abstract
Background: Plasmodium knowlesi is recognised as the main cause of human malaria in Southeast Asia. The disease is often misdiagnosed as P. falciparum or P. malariae infections by microscopy, and the disease is difficult to eliminate due to its presence in both humans and monkeys. P. knowlesi infections can rapidly cause severe disease and require prompt diagnosis and treatment. No protein biomarker exists for the rapid diagnostic test (RDT) detection of P. knowlesi infections. Plasmodium knowlesi infections can be diagnosed by PCR.
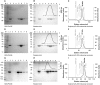
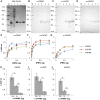
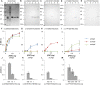
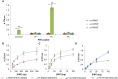
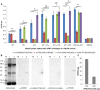
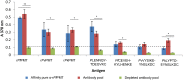
Methods and principal findings: Phosphoethanolamine-N-methyltransferase (PMT) is involved in malaria lipid biosynthesis and is not found in the human host. The P. falciparum, P. vivax and P. knowlesi PMT proteins were recombinantly expressed in BL21(DE3) Escherichia coli host cells, affinity purified and used to raise antibodies in chickens. Antibodies against each recombinant PMT protein all detected all three recombinant proteins and the native 29 kDa P. falciparum PMT protein on western blots and in ELISA. Antibodies against a PMT epitope (PLENNQYTDEGVKC) common to all three PMT orthologues detected all three proteins. Antibodies against unique peptides from each orthologue of PMT, PfCEVEHKYLHENKE, PvVYSIKEYNSLKDC, PkLYPTDEYNSLKDC detected only the parent protein in western blots and P. falciparum infected red blood cell lysates or blood lysates spiked with the respective proteins. Similar concentrations of PfPMT and the control, PfLDH, were detected in the same parasite lysate. The recombinant PfPMT protein was detected by a human anti-malaria antibody pool.
Conclusion: PMT, like the pan-specific LDH biomarker used in RDT tests, is both soluble, present at comparable concentrations in the parasite and constitutes a promising antimalarial drug target. PMT is absent from the human proteome. PMT has the potential as a biomarker for human malaria and in particular as the first P. knowlesi specific protein with diagnostic potential for the identification of a P. knowlesi infection.
Conflict of interest statement
Figures

Similar articles
-
Plasmodium glyceraldehyde-3-phosphate dehydrogenase: A potential malaria diagnostic target.Exp Parasitol. 2017 Aug;179:7-19. doi: 10.1016/j.exppara.2017.05.007. Epub 2017 May 26. Exp Parasitol. 2017. PMID: 28552792
-
Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi.Malar J. 2014 Feb 18;13:60. doi: 10.1186/1475-2875-13-60. Malar J. 2014. PMID: 24548805 Free PMC article.
-
A panel of recombinant proteins from human-infective Plasmodium species for serological surveillance.Malar J. 2020 Jan 17;19(1):31. doi: 10.1186/s12936-020-3111-5. Malar J. 2020. PMID: 31952523 Free PMC article.
-
Review of cases with the emerging fifth human malaria parasite, Plasmodium knowlesi.Clin Infect Dis. 2011 Jun;52(11):1356-62. doi: 10.1093/cid/cir180. Clin Infect Dis. 2011. PMID: 21596677 Review.
-
Plasmodium knowlesi: from Malaysia, a novel health care threat.Infez Med. 2012 Mar;20(1):5-11. Infez Med. 2012. PMID: 22475654 Review.
Cited by
-
A Dual, Systematic Approach to Malaria Diagnostic Biomarker Discovery.Clin Infect Dis. 2022 Jan 7;74(1):40-51. doi: 10.1093/cid/ciab251. Clin Infect Dis. 2022. PMID: 34718455 Free PMC article.
-
Antimalarial Properties of Isoquinoline Derivative from Streptomyces hygroscopicus subsp. Hygroscopicus: An In Silico Approach.Biomed Res Int. 2020 Jan 9;2020:6135696. doi: 10.1155/2020/6135696. eCollection 2020. Biomed Res Int. 2020. PMID: 31993450 Free PMC article.
-
Computational and experimental elucidation of Plasmodium falciparum phosphoethanolamine methyltransferase inhibitors: Pivotal drug target.PLoS One. 2019 Aug 22;14(8):e0221032. doi: 10.1371/journal.pone.0221032. eCollection 2019. PLoS One. 2019. PMID: 31437171 Free PMC article.
-
Metabolic Labeling-Based Chemoproteomics Establishes Choline Metabolites as Protein Function Modulators.ACS Chem Biol. 2022 Aug 19;17(8):2272-2283. doi: 10.1021/acschembio.2c00400. Epub 2022 Jul 8. ACS Chem Biol. 2022. PMID: 35802552 Free PMC article.
-
Plasmodium knowlesi: the game changer for malaria eradication.Malar J. 2022 May 3;21(1):140. doi: 10.1186/s12936-022-04131-8. Malar J. 2022. PMID: 35505339 Free PMC article. Review.
References
-
- Martinsen ES, Perkins SL, Schall JJ, (2008) A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. 2008; Evol. 47: 261–73. - PubMed
-
- World Health Organization (2016) World malaria report 2016. Geneva: World Health Organization.
-
- Chin W, Contacos PG, Coatney GR, Kimball HR, (1965) A Naturally Acquired Quotidian-Type Malaria in Man Transferable to Monkeys. Science. 149: 865 - PubMed
-
- Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al., (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 363: 1017–24. doi: 10.1016/S0140-6736(04)15836-4 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

