The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling
- PMID: 29505855
- PMCID: PMC6000238
- DOI: 10.1016/j.gpb.2017.10.002
The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling
Abstract
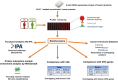
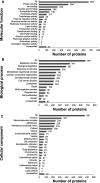
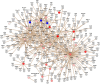
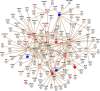
Characterization of the colon cancer immunome and its autoantibody signature from differentially-reactive antigens (DIRAGs) could provide insights into aberrant cellular mechanisms or enriched networks associated with diseases. The purpose of this study was to characterize the antibody profile of plasma samples from 32 colorectal cancer (CRC) patients and 32 controls using proteins isolated from 15,417 human cDNA expression clones on microarrays. 671 unique DIRAGs were identified and 632 were more highly reactive in CRC samples. Bioinformatics analyses reveal that compared to control samples, the immunoproteomic IgG profiling of CRC samples is mainly associated with cell death, survival, and proliferation pathways, especially proteins involved in EIF2 and mTOR signaling. Ribosomal proteins (e.g., RPL7, RPL22, and RPL27A) and CRC-related genes such as APC, AXIN1, E2F4, MSH2, PMS2, and TP53 were highly enriched. In addition, differential pathways were observed between the CRC and control samples. Furthermore, 103 DIRAGs were reported in the SEREX antigen database, demonstrating our ability to identify known and new reactive antigens. We also found an overlap of 7 antigens with 48 "CRC genes." These data indicate that immunomics profiling on protein microarrays is able to reveal the complexity of immune responses in cancerous diseases and faithfully reflects the underlying pathology.
Keywords: Autoantibody tumor biomarker; Cancer immunology; Colorectal cancer; Immunomics; Protein microarray.
Copyright © 2018 Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China. Production and hosting by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
The prostate cancer immunome: In silico functional analysis of antigenic proteins from microarray profiling with IgG.Proteomics. 2016 Apr;16(8):1204-14. doi: 10.1002/pmic.201500378. Epub 2016 Apr 4. Proteomics. 2016. PMID: 27089054
-
Autoantibody Profiling in Ulcerative Colitis: Identification of Early Immune Signatures and Disease-Associated Antigens for Improved Diagnosis and Monitoring.Int J Mol Sci. 2025 Apr 25;26(9):4086. doi: 10.3390/ijms26094086. Int J Mol Sci. 2025. PMID: 40362323 Free PMC article.
-
The pre-analytical processing of blood samples for detecting biomarkers on protein microarrays.J Immunol Methods. 2015 Mar;418:39-51. doi: 10.1016/j.jim.2015.01.009. Epub 2015 Feb 9. J Immunol Methods. 2015. PMID: 25675867
-
Functional Analysis of Autoantibody Signatures in Rheumatoid Arthritis.Molecules. 2022 Feb 21;27(4):1452. doi: 10.3390/molecules27041452. Molecules. 2022. PMID: 35209238 Free PMC article.
-
Identification of potential hub genes via bioinformatics analysis combined with experimental verification in colorectal cancer.Mol Carcinog. 2020 Apr;59(4):425-438. doi: 10.1002/mc.23165. Epub 2020 Feb 16. Mol Carcinog. 2020. PMID: 32064687
Cited by
-
Patients with Systemic Juvenile Idiopathic Arthritis (SJIA) Show Differences in Autoantibody Signatures Based on Disease Activity.Biomolecules. 2023 Sep 15;13(9):1392. doi: 10.3390/biom13091392. Biomolecules. 2023. PMID: 37759792 Free PMC article.
-
Antibody Profiling and In Silico Functional Analysis of Differentially Reactive Antibody Signatures of Glioblastomas and Meningiomas.Int J Mol Sci. 2023 Jan 11;24(2):1411. doi: 10.3390/ijms24021411. Int J Mol Sci. 2023. PMID: 36674927 Free PMC article.
-
Systemic autoinflammatory diseases.J Autoimmun. 2020 May;109:102421. doi: 10.1016/j.jaut.2020.102421. Epub 2020 Feb 1. J Autoimmun. 2020. PMID: 32019685 Free PMC article. Review.
-
A Master Autoantigen-ome Links Alternative Splicing, Female Predilection, and COVID-19 to Autoimmune Diseases.bioRxiv [Preprint]. 2021 Aug 4:2021.07.30.454526. doi: 10.1101/2021.07.30.454526. bioRxiv. 2021. Update in: J Transl Autoimmun. 2022;5:100147. doi: 10.1016/j.jtauto.2022.100147. PMID: 34373855 Free PMC article. Updated. Preprint.
-
Tracking the Antibody Immunome in Sporadic Colorectal Cancer by Using Antigen Self-Assembled Protein Arrays.Cancers (Basel). 2021 May 31;13(11):2718. doi: 10.3390/cancers13112718. Cancers (Basel). 2021. PMID: 34072782 Free PMC article.
References
-
- Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Edwards B.K., Ward E., Kohler B.A., Eheman C., Zauber A.G., Anderson R.N. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–573. - PMC - PubMed
-
- Luna Coronell J.A., Syed P., Sergelen K., Gyurján I., Weinhäusel A. The current status of cancer biomarker research using tumour-associated antigens for minimal invasive and early cancer diagnostics. J Proteomics. 2012;76:102–115. - PubMed
-
- Barderas R., Babel I., Díaz-Uriarte R., Moreno V., Suárez A., Bonilla F. An optimized predictor panel for colorectal cancer diagnosis based on the combination of tumor-associated antigens obtained from protein and phage microarrays. J Proteomics. 2012;75:4647–4655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

