Chemotaxis Allows Bacteria To Overcome Host-Generated Reactive Oxygen Species That Constrain Gland Colonization
- PMID: 29507083
- PMCID: PMC5913845
- DOI: 10.1128/IAI.00878-17
Chemotaxis Allows Bacteria To Overcome Host-Generated Reactive Oxygen Species That Constrain Gland Colonization
Abstract
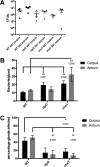
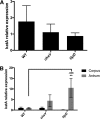
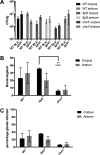
The epithelial layer of the gastrointestinal tract contains invaginations, called glands or crypts, which are colonized by symbiotic and pathogenic microorganisms and may function as designated niches for certain species. Factors that control gland colonization are poorly understood, but bacterial chemotaxis aids occupation of these sites. We report here that a Helicobacter pylori cytoplasmic chemoreceptor, TlpD, is required for gland colonization in the stomach. tlpD mutants demonstrate gland colonization defects characterized by a reduction in the percentage of glands colonized but not in the number of bacteria per gland. Consistent with TlpD's reported role in reactive oxygen species (ROS) avoidance, tlpD mutants showed hallmarks of exposure to high ROS. To assess the role of host-generated ROS in TlpD-dependent gland colonization, we utilized mice that lack either the ability to generate epithelial hydrogen peroxide or immune cell superoxide. tlpD gland colonization defects were rescued to wild-type H. pylori levels in both of these mutants. These results suggest that multiple types of innate immune-generated ROS production limit gland colonization and that bacteria have evolved specific mechanisms to sense and direct their motility in response to this signal and thus spread throughout tissue.
Keywords: MCP; biogeography; chemoreceptors; crypts; glands; reactive oxygen species; signal transduction; stomach.
Copyright © 2018 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

