Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats
- PMID: 29507093
- PMCID: PMC5936814
- DOI: 10.1074/jbc.RA117.000347
Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats
Erratum in
-
Correction: Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats.J Biol Chem. 2019 May 24;294(21):8348. doi: 10.1074/jbc.AAC119.009120. J Biol Chem. 2019. PMID: 31127060 Free PMC article. No abstract available.
Abstract
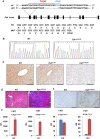
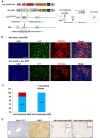
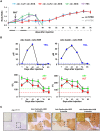
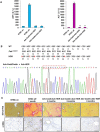
Hereditary tyrosinemia type I (HTI) is a metabolic genetic disorder caused by mutation of fumarylacetoacetate hydrolase (FAH). Because of the accumulation of toxic metabolites, HTI causes severe liver cirrhosis, liver failure, and even hepatocellular carcinoma. HTI is an ideal model for gene therapy, and several strategies have been shown to ameliorate HTI symptoms in animal models. Although CRISPR/Cas9-mediated genome editing is able to correct the Fah mutation in mouse models, WT Cas9 induces numerous undesired mutations that have raised safety concerns for clinical applications. To develop a new method for gene correction with high fidelity, we generated a Fah mutant rat model to investigate whether Cas9 nickase (Cas9n)-mediated genome editing can efficiently correct the Fah First, we confirmed that Cas9n rarely induces indels in both on-target and off-target sites in cell lines. Using WT Cas9 as a positive control, we delivered Cas9n and the repair donor template/single guide (sg)RNA through adenoviral vectors into HTI rats. Analyses of the initial genome editing efficiency indicated that only WT Cas9 but not Cas9n causes indels at the on-target site in the liver tissue. After receiving either Cas9n or WT Cas9-mediated gene correction therapy, HTI rats gained weight steadily and survived. Fah-expressing hepatocytes occupied over 95% of the liver tissue 9 months after the treatment. Moreover, CRISPR/Cas9-mediated gene therapy prevented the progression of liver cirrhosis, a phenotype that could not be recapitulated in the HTI mouse model. These results strongly suggest that Cas9n-mediated genome editing is a valuable and safe gene therapy strategy for this genetic disease.
Keywords: CRISPR/Cas; Cas9 nickase; fibrosis; gene editing; gene therapy; genetic disease; genome editing; hereditary tyrosinemia; liver.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Curative Ex Vivo Hepatocyte-Directed Gene Editing in a Mouse Model of Hereditary Tyrosinemia Type 1.Hum Gene Ther. 2018 Nov;29(11):1315-1326. doi: 10.1089/hum.2017.252. Epub 2018 Jun 22. Hum Gene Ther. 2018. PMID: 29764210 Free PMC article.
-
Efficient liver repopulation of transplanted hepatocyte prevents cirrhosis in a rat model of hereditary tyrosinemia type I.Sci Rep. 2016 Aug 11;6:31460. doi: 10.1038/srep31460. Sci Rep. 2016. PMID: 27510266 Free PMC article.
-
Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype.Nat Biotechnol. 2014 Jun;32(6):551-3. doi: 10.1038/nbt.2884. Epub 2014 Mar 30. Nat Biotechnol. 2014. PMID: 24681508 Free PMC article.
-
Nuclease-Mediated Gene Therapies for Inherited Metabolic Diseases of the Liver.Yale J Biol Med. 2017 Dec 19;90(4):553-566. eCollection 2017 Dec. Yale J Biol Med. 2017. PMID: 29259521 Free PMC article. Review.
-
CRISPR/Cas9: targeted genome editing for the treatment of hereditary hearing loss.J Appl Genet. 2020 Feb;61(1):51-65. doi: 10.1007/s13353-019-00535-6. Epub 2020 Jan 7. J Appl Genet. 2020. PMID: 31912450 Review.
Cited by
-
Ex Vivo Hepatocyte Reprograming Promotes Homology-Directed DNA Repair to Correct Metabolic Disease in Mice After Transplantation.Hepatol Commun. 2019 Feb 15;3(4):558-573. doi: 10.1002/hep4.1315. eCollection 2019 Apr. Hepatol Commun. 2019. PMID: 30976745 Free PMC article.
-
CRISPR-Cas9 System for Genome Engineering of Photosynthetic Microalgae.Mol Biotechnol. 2019 Aug;61(8):541-561. doi: 10.1007/s12033-019-00185-3. Mol Biotechnol. 2019. PMID: 31140149 Review.
-
Advanced delivery systems for gene editing: A comprehensive review from the GenE-HumDi COST Action Working Group.Mol Ther Nucleic Acids. 2025 Jan 17;36(1):102457. doi: 10.1016/j.omtn.2025.102457. eCollection 2025 Mar 11. Mol Ther Nucleic Acids. 2025. PMID: 39991472 Free PMC article. Review.
-
Recent advances in therapeutic gene-editing technologies.Mol Ther. 2025 Jun 4;33(6):2619-2644. doi: 10.1016/j.ymthe.2025.03.026. Epub 2025 Mar 20. Mol Ther. 2025. PMID: 40119516 Review.
-
Knockdown of lactate dehydrogenase by adeno-associated virus-delivered CRISPR/Cas9 system alleviates primary hyperoxaluria type 1.Clin Transl Med. 2020 Dec;10(8):e261. doi: 10.1002/ctm2.261. Clin Transl Med. 2020. PMID: 33377632 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

