Rap1, Canoe and Mbt cooperate with Bazooka to promote zonula adherens assembly in the fly photoreceptor
- PMID: 29507112
- PMCID: PMC5897711
- DOI: 10.1242/jcs.207779
Rap1, Canoe and Mbt cooperate with Bazooka to promote zonula adherens assembly in the fly photoreceptor
Abstract
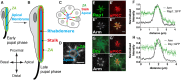
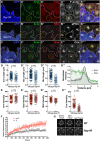
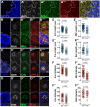
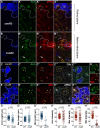
In Drosophila epithelial cells, apical exclusion of Bazooka (the Drosophila Par3 protein) defines the position of the zonula adherens (ZA), which demarcates the apical and lateral membrane and allows cells to assemble into sheets. Here, we show that the small GTPase Rap1, its effector Canoe (Cno) and the Cdc42 effector kinase Mushroom bodies tiny (Mbt), converge in regulating epithelial morphogenesis by coupling stabilization of the adherens junction (AJ) protein E-Cadherin and Bazooka retention at the ZA. Furthermore, our results show that the localization of Rap1, Cno and Mbt at the ZA is interdependent, indicating that their functions during ZA morphogenesis are interlinked. In this context, we find the Rap1-GEF Dizzy is enriched at the ZA and our results suggest that it promotes Rap1 activity during ZA morphogenesis. Altogether, we propose the Dizzy, Rap1 and Cno pathway and Mbt converge in regulating the interface between Bazooka and AJ material to promote ZA morphogenesis.
Keywords: AFDN; Bazooka; Epithelial polarity; Pak4; Par3; Photoreceptor; Rap1; Zonula adherens.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

