dCas9-targeted locus-specific protein isolation method identifies histone gene regulators
- PMID: 29507191
- PMCID: PMC5866577
- DOI: 10.1073/pnas.1718844115
dCas9-targeted locus-specific protein isolation method identifies histone gene regulators
Abstract
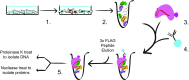
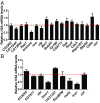
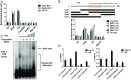
Eukaryotic gene regulation is a complex process, often coordinated by the action of tens to hundreds of proteins. Although previous biochemical studies have identified many components of the basal machinery and various ancillary factors involved in gene regulation, numerous gene-specific regulators remain undiscovered. To comprehensively survey the proteome directing gene expression at a specific genomic locus of interest, we developed an in vitro nuclease-deficient Cas9 (dCas9)-targeted chromatin-based purification strategy, called "CLASP" (Cas9 locus-associated proteome), to identify and functionally test associated gene-regulatory factors. Our CLASP method, coupled to mass spectrometry and functional screens, can be efficiently adapted for isolating associated regulatory factors in an unbiased manner targeting multiple genomic loci across different cell types. Here, we applied our method to isolate the Drosophila melanogaster histone cluster in S2 cells to identify several factors including Vig and Vig2, two proteins that bind and regulate core histone H2A and H3 mRNA via interaction with their 3' UTRs.
Keywords: CRISPR/Cas9; gene expression; histone regulation; reverse ChIP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Nucleic Acid-Dependent Conformational Changes in CRISPR-Cas9 Revealed by Site-Directed Spin Labeling.Cell Biochem Biophys. 2017 Jun;75(2):203-210. doi: 10.1007/s12013-016-0738-5. Epub 2016 Jun 24. Cell Biochem Biophys. 2017. PMID: 27342128 Free PMC article.
-
Gene activation by dCas9-CBP and the SAM system differ in target preference.Sci Rep. 2019 Dec 2;9(1):18104. doi: 10.1038/s41598-019-54179-x. Sci Rep. 2019. PMID: 31792240 Free PMC article.
-
CRISPR/Cas9 in Genome Editing and Beyond.Annu Rev Biochem. 2016 Jun 2;85:227-64. doi: 10.1146/annurev-biochem-060815-014607. Epub 2016 Apr 25. Annu Rev Biochem. 2016. PMID: 27145843 Review.
-
Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila.Genetics. 2014 Apr;196(4):961-71. doi: 10.1534/genetics.113.160713. Epub 2014 Jan 29. Genetics. 2014. PMID: 24478335 Free PMC article.
-
[CRISPR/CAS9, the King of Genome Editing Tools].Mol Biol (Mosk). 2017 Jul-Aug;51(4):582-594. doi: 10.7868/S0026898417040036. Mol Biol (Mosk). 2017. PMID: 28900076 Review. Russian.
Cited by
-
The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation.Genome Biol. 2020 Aug 6;21(1):195. doi: 10.1186/s13059-020-02115-y. Genome Biol. 2020. PMID: 32762776 Free PMC article.
-
CRISPR/Cas9 Landscape: Current State and Future Perspectives.Int J Mol Sci. 2023 Nov 8;24(22):16077. doi: 10.3390/ijms242216077. Int J Mol Sci. 2023. PMID: 38003266 Free PMC article. Review.
-
Cardiac enhancers: Gateway to the regulatory mechanisms of heart regeneration.Semin Cell Dev Biol. 2025 Jun;170:103610. doi: 10.1016/j.semcdb.2025.103610. Epub 2025 Apr 10. Semin Cell Dev Biol. 2025. PMID: 40215762 Review.
-
Applications of CRISPR-Cas Technologies to Proteomics.Genes (Basel). 2021 Nov 12;12(11):1790. doi: 10.3390/genes12111790. Genes (Basel). 2021. PMID: 34828396 Free PMC article. Review.
-
Salinity-responsive histone PTMs identified in the gills and gonads of Mozambique tilapia (Oreochromis mossambicus).BMC Genomics. 2024 Jun 11;25(1):586. doi: 10.1186/s12864-024-10471-3. BMC Genomics. 2024. PMID: 38862901 Free PMC article.
References
-
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
-
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. - PubMed
-
- Strausbaugh LD, Weinberg ES. Polymorphism and stability in the histone gene cluster of Drosophila melanogaster. Chromosoma. 1982;85:489–505. - PubMed
-
- Marzluff WF, Duronio RJ. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

