Mechanism of the G-protein mimetic nanobody binding to a muscarinic G-protein-coupled receptor
- PMID: 29507218
- PMCID: PMC5866610
- DOI: 10.1073/pnas.1800756115
Mechanism of the G-protein mimetic nanobody binding to a muscarinic G-protein-coupled receptor
Abstract
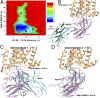
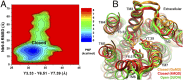
Protein-protein binding is key in cellular signaling processes. Molecular dynamics (MD) simulations of protein-protein binding, however, are challenging due to limited timescales. In particular, binding of the medically important G-protein-coupled receptors (GPCRs) with intracellular signaling proteins has not been simulated with MD to date. Here, we report a successful simulation of the binding of a G-protein mimetic nanobody to the M2 muscarinic GPCR using the robust Gaussian accelerated MD (GaMD) method. Through long-timescale GaMD simulations over 4,500 ns, the nanobody was observed to bind the receptor intracellular G-protein-coupling site, with a minimum rmsd of 2.48 Å in the nanobody core domain compared with the X-ray structure. Binding of the nanobody allosterically closed the orthosteric ligand-binding pocket, being consistent with the recent experimental finding. In the absence of nanobody binding, the receptor orthosteric pocket sampled open and fully open conformations. The GaMD simulations revealed two low-energy intermediate states during nanobody binding to the M2 receptor. The flexible receptor intracellular loops contribute remarkable electrostatic, polar, and hydrophobic residue interactions in recognition and binding of the nanobody. These simulations provided important insights into the mechanism of GPCR-nanobody binding and demonstrated the applicability of GaMD in modeling dynamic protein-protein interactions.
Keywords: GPCR signaling; biomolecular recognition; enhanced sampling; pathways; protein binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Spalding TA, Burstein ES. Constitutive activity of muscarinic acetylcholine receptors. J Recept Signal Transduct Res. 2006;26:61–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

