Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteri in gut microbe-host interactions
- PMID: 29507249
- PMCID: PMC5866549
- DOI: 10.1073/pnas.1715016115
Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteri in gut microbe-host interactions
Abstract
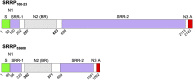
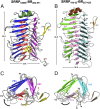
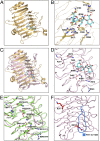
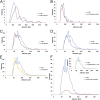
Lactobacillus reuteri, a Gram-positive bacterial species inhabiting the gastrointestinal tract of vertebrates, displays remarkable host adaptation. Previous mutational analyses of rodent strain L. reuteri 100-23C identified a gene encoding a predicted surface-exposed serine-rich repeat protein (SRRP100-23) that was vital for L. reuteri biofilm formation in mice. SRRPs have emerged as an important group of surface proteins on many pathogens, but no structural information is available in commensal bacteria. Here we report the 2.00-Å and 1.92-Å crystal structures of the binding regions (BRs) of SRRP100-23 and SRRP53608 from L. reuteri ATCC 53608, revealing a unique β-solenoid fold in this important adhesin family. SRRP53608-BR bound to host epithelial cells and DNA at neutral pH and recognized polygalacturonic acid (PGA), rhamnogalacturonan I, or chondroitin sulfate A at acidic pH. Mutagenesis confirmed the role of the BR putative binding site in the interaction of SRRP53608-BR with PGA. Long molecular dynamics simulations showed that SRRP53608-BR undergoes a pH-dependent conformational change. Together, these findings provide mechanistic insights into the role of SRRPs in host-microbe interactions and open avenues of research into the use of biofilm-forming probiotics against clinically important pathogens.
Keywords: Lactobacillus reuteri; SRRP; adhesin; biofilm; mucin.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Serine-rich repeat proteins: well-known yet little-understood bacterial adhesins.J Bacteriol. 2024 Jan 25;206(1):e0024123. doi: 10.1128/jb.00241-23. Epub 2023 Nov 17. J Bacteriol. 2024. PMID: 37975670 Free PMC article. Review.
-
Serine-rich repeat protein adhesins from Lactobacillus reuteri display strain specific glycosylation profiles.Glycobiology. 2019 Jan 1;29(1):45-58. doi: 10.1093/glycob/cwy100. Glycobiology. 2019. PMID: 30371779 Free PMC article.
-
Structural and molecular insights into novel surface-exposed mucus adhesins from Lactobacillus reuteri human strains.Mol Microbiol. 2014 May;92(3):543-56. doi: 10.1111/mmi.12574. Epub 2014 Apr 2. Mol Microbiol. 2014. PMID: 24593252
-
Role of Lactobacillus reuteri cell and mucus-binding protein A (CmbA) in adhesion to intestinal epithelial cells and mucus in vitro.Microbiology (Reading). 2014 Apr;160(Pt 4):671-681. doi: 10.1099/mic.0.073551-0. Epub 2014 Jan 28. Microbiology (Reading). 2014. PMID: 24473252 Free PMC article.
-
Serine-rich repeat proteins from gut microbes.Gut Microbes. 2020;11(1):102-117. doi: 10.1080/19490976.2019.1602428. Epub 2019 Apr 29. Gut Microbes. 2020. PMID: 31035824 Free PMC article. Review.
Cited by
-
Serine-rich repeat proteins: well-known yet little-understood bacterial adhesins.J Bacteriol. 2024 Jan 25;206(1):e0024123. doi: 10.1128/jb.00241-23. Epub 2023 Nov 17. J Bacteriol. 2024. PMID: 37975670 Free PMC article. Review.
-
A conserved bacterial genetic basis for commensal-host specificity.Science. 2024 Dec 6;386(6726):1117-1122. doi: 10.1126/science.adp7748. Epub 2024 Dec 5. Science. 2024. PMID: 39636981
-
Lactobacilli and human dental caries: more than mechanical retention.Microbiology (Reading). 2022 Jun;168(6):001196. doi: 10.1099/mic.0.001196. Microbiology (Reading). 2022. PMID: 35671222 Free PMC article.
-
Impact of probiotic Limosilactobacillus reuteri DSM 17938 on amino acid metabolism in the healthy newborn mouse.Amino Acids. 2022 Oct;54(10):1383-1401. doi: 10.1007/s00726-022-03165-1. Epub 2022 May 10. Amino Acids. 2022. PMID: 35536363 Free PMC article.
-
The role of the glycome in symbiotic host-microbe interactions.Glycobiology. 2023 Dec 30;33(12):1106-1116. doi: 10.1093/glycob/cwad073. Glycobiology. 2023. PMID: 37741057 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- BBS/E/F/00044452/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K019554/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT106121MA/WT_/Wellcome Trust/United Kingdom
- BBS/E/F/000PR10356/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004529/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P010660/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BBS/E/F/000PR10355/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

