A lanthipeptide library used to identify a protein-protein interaction inhibitor
- PMID: 29507389
- PMCID: PMC5866752
- DOI: 10.1038/s41589-018-0008-5
A lanthipeptide library used to identify a protein-protein interaction inhibitor
Abstract
In this article we describe the production and screening of a genetically encoded library of 106 lanthipeptides in Escherichia coli using the substrate-tolerant lanthipeptide synthetase ProcM. This plasmid-encoded library was combined with a bacterial reverse two-hybrid system for the interaction of the HIV p6 protein with the UEV domain of the human TSG101 protein, which is a critical protein-protein interaction for HIV budding from infected cells. Using this approach, we identified an inhibitor of this interaction from the lanthipeptide library, whose activity was verified in vitro and in cell-based virus-like particle-budding assays. Given the variety of lanthipeptide backbone scaffolds that may be produced with ProcM, this method may be used for the generation of genetically encoded libraries of natural product-like lanthipeptides containing substantial structural diversity. Such libraries may be combined with any cell-based assay to identify lanthipeptides with new biological activities.
Figures


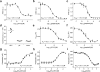
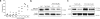
References
-
- Marsault E, Peterson ML. Macrocycles are great cycles: applications, opportunities, and challenges of synthetic macrocycles in drug discovery. J Med Chem. 2011;54:1961–2004. - PubMed
-
- Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery-an underexploited structural class. Nat Rev Drug Discov. 2008;7:608–624. - PubMed
-
- Lennard KR, Tavassoli A. Peptides come round: using SICLOPPS libraries for early stage drug discovery. Chemistry. 2014;20:10608–10614. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

