Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons
- PMID: 29507411
- PMCID: PMC6503322
- DOI: 10.1038/s41593-018-0091-7
Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons
Abstract
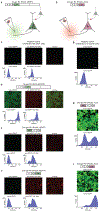
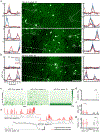
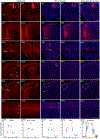
Recombinant rabies viral vectors have proven useful for applications including retrograde targeting of projection neurons and monosynaptic tracing, but their cytotoxicity has limited their use to short-term experiments. Here we introduce a new class of double-deletion-mutant rabies viral vectors that left transduced cells alive and healthy indefinitely. Deletion of the viral polymerase gene abolished cytotoxicity and reduced transgene expression to trace levels but left vectors still able to retrogradely infect projection neurons and express recombinases, allowing downstream expression of other transgene products such as fluorophores and calcium indicators. The morphology of retrogradely targeted cells appeared unperturbed at 1 year postinjection. Whole-cell patch-clamp recordings showed no physiological abnormalities at 8 weeks. Longitudinal two-photon structural and functional imaging in vivo, tracking thousands of individual neurons for up to 4 months, showed that transduced neurons did not die but retained stable visual response properties even at the longest time points imaged.
Conflict of interest statement
Competing financial interests statement
The authors have no competing financial interests.
Figures

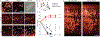

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

