Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma
- PMID: 29507645
- PMCID: PMC5833129
- DOI: 10.1186/s13148-018-0462-7
Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma
Abstract
Background: Hepatoblastoma (HB) is the most common liver tumor of childhood and occurs predominantly within the first 3 years of life. In accordance to its early manifestation, HB has been described to display an extremely low mutation rate. As substitute, epigenetic modifiers seem to play an exceptional role in its tumorigenesis, which holds promise to develop targeted therapies and establish biomarkers for patient risk stratification.
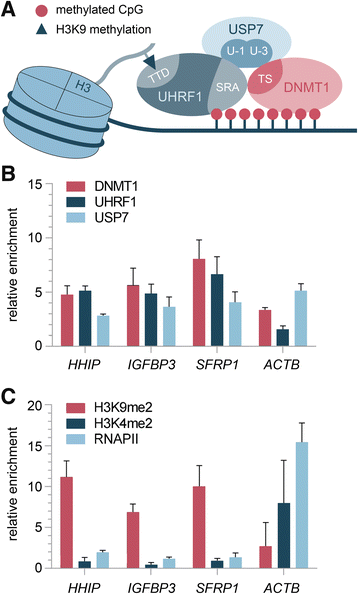
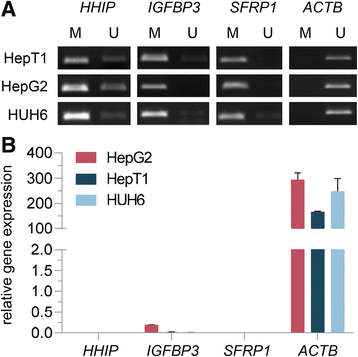
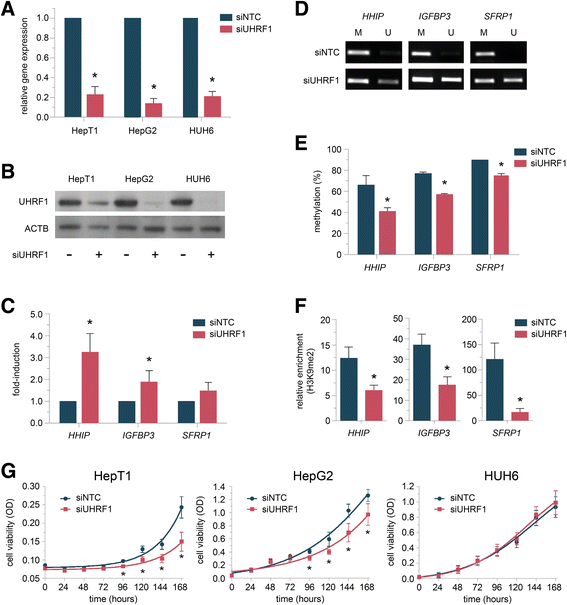
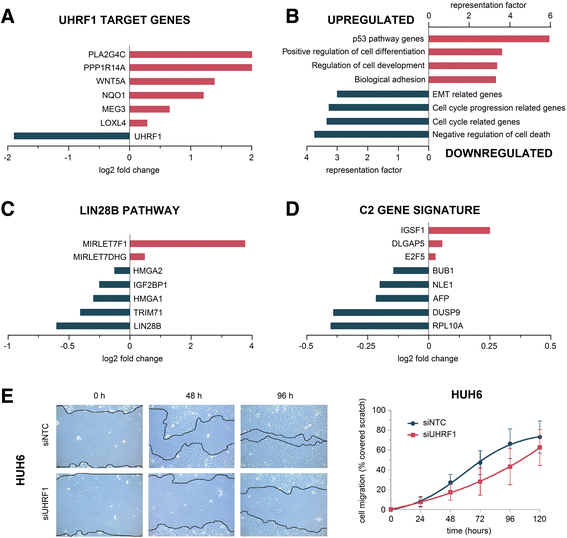
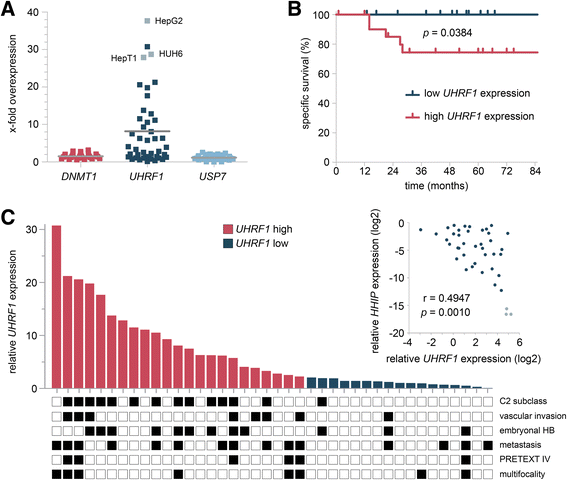
Results: We examined the role of a newly described protein complex consisting of three epigenetic regulators, namely E3 ubiquitin-like containing PHD and RING finger domain 1 (UHRF1), ubiquitin-specific-processing protease 7 (USP7), and DNA methyltransferase 1 (DNMT1), in HB. We found the complex to be located on the promoter regions of the pivotal HB-associated tumor suppressor genes (TSGs) HHIP, IGFBP3, and SFRP1 in HB cells, thereby leading to strong repression through DNA methylation and histone modifications. Consequently, knockdown of UHRF1 led to DNA demethylation and loss of the repressive H3K9me2 histone mark at the TSG loci with their subsequent transcriptional reactivation. The observed growth impairment of HB cells upon UHRF1 knockdown could be attributed to reduced expression of genes involved in cell cycle progression, negative regulation of cell death, LIN28B signaling, and the adverse 16-gene signature, as revealed by global RNA sequencing. Clinically, overexpression of UHRF1 in primary tumor tissues was significantly associated with poor survival and the prognostic high-risk 16-gene signature.
Conclusion: These findings suggest that UHRF1 is critical for aberrant TSG silencing and sustained growth signaling in HB and that UHRF1 overexpression levels might serve as a prognostic biomarker and potential molecular target for HB patients.
Keywords: Biomarker; DNA methylation; DNMT1; Epigenetic silencing; Hepatoblastoma; Histone methylation; Tumor suppressor genes; UHRF1; USP7.
Conflict of interest statement
Written informed consent was obtained from each patient, and the study protocol was approved by the Committee of Ethics of the Ludwig Maximilian University of Munich (no. 431-11).Not applicableThe authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Meyers RL, Maibach R, Hiyama E, Haberle B, Krailo M, Rangaswami A, Aronson DC, Malogolowkin MH, Perilongo G, von Schweinitz D, et al. Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children’s Hepatic tumors International Collaboration. Lancet Oncol. 2017;18:122–131. doi: 10.1016/S1470-2045(16)30598-8. - DOI - PMC - PubMed
-
- Zsiros J, Maibach R, Shafford E, Brugieres L, Brock P, Czauderna P, Roebuck D, Childs M, Zimmermann A, Laithier V, et al. Successful treatment of childhood high-risk hepatoblastoma with dose-intensive multiagent chemotherapy and surgery: final results of the SIOPEL-3HR study. J Clin Oncol. 2010;28:2584–2590. doi: 10.1200/JCO.2009.22.4857. - DOI - PubMed
-
- Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, Goga A, Balakrishnan A, Semeraro M, Gresh L, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

