New localization and function of calpain-2 in nucleoli of colorectal cancer cells in ribosomal biogenesis: effect of KRAS status
- PMID: 29507677
- PMCID: PMC5823616
- DOI: 10.18632/oncotarget.23888
New localization and function of calpain-2 in nucleoli of colorectal cancer cells in ribosomal biogenesis: effect of KRAS status
Erratum in
-
Correction: New localization and function of calpain-2 in nucleoli of colorectal cancer cells in ribosomal biogenesis: effect of KRAS status.Oncotarget. 2018 Apr 6;9(26):18664. doi: 10.18632/oncotarget.25142. eCollection 2018 Apr 6. Oncotarget. 2018. PMID: 29722369 Free PMC article.
Abstract
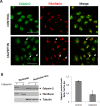
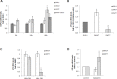
Calpain-2 belongs to a family of pleiotropic Cys-proteases with modulatory rather than degradative functions. Calpain (CAPN) overexpression has been controversially correlated with poor prognosis in several cancer types, including colorectal carcinoma (CRC). However, the mechanisms of substrate-recognition, calpain-2 regulation/deregulation and specific functions in CRC remain elusive. Herein, calpain subcellular distribution was studied as a key event for substrate-recognition and consequently, for calpain-mediated function. We describe a new localization for calpain-2 in the nucleoli of CRC cells. Calpain-2 nucleolar distribution resulted dependent on its enzymatic activity and on the mutational status of KRAS. In KRASWT/- cells serum-starvation induced CAPN2 expression, nucleolar accumulation and increased binding to the rDNA-core promoter and intergenic spacer (IGS), concomitant with a reduction in pre-rRNA levels. Depletion of calpain-2 by specific siRNA prevented pre-rRNA down-regulation after serum removal. Conversely, ribosomal biogenesis proceeded in the absence of serum in unresponsive KRASG13D/- cells whose CAPN2 expression, nucleolar localization and rDNA-occupancy remained unchanged during the time-course of serum starvation. We propose here that nucleolar calpain-2 might be a KRAS-dependent sensor to repress ribosomal biogenesis in growth limiting conditions. Under constitutive activation of the pathway commonly found in CRC, calpain-2 is deregulated and tumor cells become insensitive to the extracellular microenvironment.
Keywords: colorectal cancer; nucleolar calpain-2; pre-rRNA; serum starvation; subcellular localization.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures






Similar articles
-
Correction: New localization and function of calpain-2 in nucleoli of colorectal cancer cells in ribosomal biogenesis: effect of KRAS status.Oncotarget. 2018 Apr 6;9(26):18664. doi: 10.18632/oncotarget.25142. eCollection 2018 Apr 6. Oncotarget. 2018. PMID: 29722369 Free PMC article.
-
EGF-Induced Acetylation of Heterogeneous Nuclear Ribonucleoproteins Is Dependent on KRAS Mutational Status in Colorectal Cancer Cells.PLoS One. 2015 Jun 25;10(6):e0130543. doi: 10.1371/journal.pone.0130543. eCollection 2015. PLoS One. 2015. PMID: 26110767 Free PMC article.
-
Isoform-specific function of calpains in cell adhesion disruption: studies in postlactational mammary gland and breast cancer.Biochem J. 2016 Sep 15;473(18):2893-909. doi: 10.1042/BCJ20160198. Epub 2016 Jul 11. Biochem J. 2016. PMID: 27402795
-
Post-transcriptional regulation of ribosomal protein genes during serum starvation in Entamoeba histolytica.Mol Biochem Parasitol. 2015 Jun;201(2):146-52. doi: 10.1016/j.molbiopara.2015.07.006. Epub 2015 Aug 3. Mol Biochem Parasitol. 2015. PMID: 26247142
-
The functional organization of the nucleolus in proliferating plant cells.Eur J Histochem. 2000;44(2):117-31. Eur J Histochem. 2000. PMID: 10968360 Review.
Cited by
-
Ribosome Biogenesis Alterations in Colorectal Cancer.Cells. 2020 Oct 27;9(11):2361. doi: 10.3390/cells9112361. Cells. 2020. PMID: 33120992 Free PMC article. Review.
-
Targeting Protein Synthesis in Colorectal Cancer.Cancers (Basel). 2020 May 21;12(5):1298. doi: 10.3390/cancers12051298. Cancers (Basel). 2020. PMID: 32455578 Free PMC article. Review.
-
Calpains, the proteases of two faces controlling the epithelial homeostasis in mammary gland.Front Cell Dev Biol. 2023 Sep 19;11:1249317. doi: 10.3389/fcell.2023.1249317. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37795261 Free PMC article. Review.
-
Cleavage and activation of LIM kinase 1 as a novel mechanism for calpain 2-mediated regulation of nuclear dynamics.Sci Rep. 2021 Aug 11;11(1):16339. doi: 10.1038/s41598-021-95797-8. Sci Rep. 2021. PMID: 34381117 Free PMC article.
References
-
- Sorimachi H, Hata S, Ono Y. Impact of genetic insights into calpain biology. J Biochem. 2011;150:23–37. - PubMed
-
- Campbell RL, Davies PL. Structure-function relationships in calpains. Biochem J. 2012;447:335–51. - PubMed
-
- Ono Y, Saido TC, Sorimachi H. Calpain research for drug discovery: challenges and potential. Nat Rev Drug Discov. 2016;15:854–876. - PubMed
-
- Sorimachi H, Mamitsuka H, Ono Y. Understanding the substrate specificity of conventional calpains. Biol Chem. 2012;393:853–71. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

