Multi-chaperone function modulation and association with cytoskeletal proteins are key features of the function of AIP in the pituitary gland
- PMID: 29507682
- PMCID: PMC5823669
- DOI: 10.18632/oncotarget.24183
Multi-chaperone function modulation and association with cytoskeletal proteins are key features of the function of AIP in the pituitary gland
Abstract
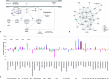
Despite the well-recognized role of loss-of-function mutations of the aryl hydrocarbon receptor interacting protein gene (AIP) predisposing to pituitary adenomas, the pituitary-specific function of this tumor suppressor remains an enigma. To determine the repertoire of interacting partners for the AIP protein in somatotroph cells, wild-type and variant AIP proteins were used for pull-down/quantitative mass spectrometry experiments against lysates of rat somatotropinoma-derived cells; relevant findings were validated by co-immunoprecipitation and co-localization. Global gene expression was studied in AIP mutation positive and negative pituitary adenomas via RNA microarrays. Direct interaction with AIP was confirmed for three known and six novel partner proteins. Novel interactions with HSPA5 and HSPA9, together with known interactions with HSP90AA1, HSP90AB1 and HSPA8, indicate that the function/stability of multiple chaperone client proteins could be perturbed by a deficient AIP co-chaperone function. Interactions with TUBB, TUBB2A, NME1 and SOD1 were also identified. The AIP variants p.R304* and p.R304Q showed impaired interactions with HSPA8, HSP90AB1, NME1 and SOD1; p.R304* also displayed reduced binding to TUBB and TUBB2A, and AIP-mutated tumors showed reduced TUBB2A expression. Our findings suggest that cytoskeletal organization, cell motility/adhesion, as well as oxidative stress responses, are functions that are likely to be involved in the tumor suppressor activity of AIP.
Keywords: AIP; FIPA; acromegaly; co-chaperone; quantitative mass spectrometry.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have nothing to disclose.
Figures


References
-
- Chahal HS, Trivellin G, Leontiou CA, Alband N, Fowkes RC, Tahir A, Igreja SC, Chapple JP, Jordan S, Lupp A, Schulz S, Ansorge O, Karavitaki N, et al. Somatostatin analogs modulate AIP in somatotroph adenomas: the role of the ZAC1 pathway. J Clin Endocrinol Metab. 2012;97:E1411–20. https://doi.org/10.1210/jc.2012-1111. - DOI - PubMed
-
- Formosa R, Xuereb-Anastasi A, Vassallo J. Aip regulates cAMP signalling and GH secretion in GH3 cells. Endocr Relat Cancer. 2013;20:495–505. https://doi.org/10.1530/ERC-13-0043. - DOI - PubMed
-
- Tuominen I, Heliovaara E, Raitila A, Rautiainen MR, Mehine M, Katainen R, Donner I, Aittomaki V, Lehtonen HJ, Ahlsten M, Kivipelto L, Schalin-Jantti C, Arola J, et al. AIP inactivation leads to pituitary tumorigenesis through defective Galpha-cAMP signaling. Oncogene. 2015;34:1174–84. https://doi.org/10.1038/onc.2014.50. - DOI - PubMed
-
- Vierimaa O, Georgitsi M, Lehtonen R, Vahteristo P, Kokko A, Raitila A, Tuppurainen K, Ebeling TM, Salmela PI, Paschke R, Gundogdu S, De Menis E, Makinen MJ, et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312:1228–30. https://doi.org/10.1126/science.1126100. - DOI - PubMed
-
- Daly AF, Tichomirowa MA, Petrossians P, Heliovaara E, Jaffrain-Rea ML, Barlier A, Naves LA, Ebeling T, Karhu A, Raappana A, Cazabat L, De Menis E, Montanana CF, et al. Clinical characteristics and therapeutic responses in patients with germ-line AIP mutations and pituitary adenomas: an international collaborative study. J Clin Endocrinol Metab. 2010;95:E373–83. https://doi.org/10.1210/jc.2009-2556. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

