Copper/MYC/CTR1 interplay: a dangerous relationship in hepatocellular carcinoma
- PMID: 29507693
- PMCID: PMC5823635
- DOI: 10.18632/oncotarget.24282
Copper/MYC/CTR1 interplay: a dangerous relationship in hepatocellular carcinoma
Abstract
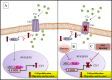
Free serum copper correlates with tumor incidence and progression of human cancers, including hepatocellular carcinoma (HCC). Copper extracellular uptake is provided by the transporter CTR1, whose expression is regulated to avoid excessive intracellular copper entry. Inadequate copper serum concentration is involved in the pathogenesis of Non Alcoholic Fatty Liver Disease (NAFLD), which is becoming a major cause of liver damage progression and HCC incidence. Finally, MYC is over-expressed in most of HCCs and is a critical regulator of cellular growth, tumor invasion and metastasis. The purpose of our study was to understand if higher serum copper concentrations might be involved in the progression of NAFLD-cirrhosis toward-HCC. We investigated whether high exogenous copper levels sensitize liver cells to transformation and if it exists an interplay between copper-related proteins and MYC oncogene. NAFLD-cirrhotic patients were characterized by a statistical significant enhancement of serum copper levels, even more evident in HCC patients. We demonstrated that high extracellular copper concentrations increase cell growth, migration, and invasion of liver cancer cells by modulating MYC/CTR1 axis. We highlighted that MYC binds a specific region of the CTR1 promoter, regulating its transcription. Accordingly, CTR1 and MYC proteins expression were progressively up-regulated in liver tissues from NAFLD-cirrhotic to HCC patients. This work provides novel insights on the molecular mechanisms by which copper may favor the progression from cirrhosis to cancer. The Cu/MYC/CTR1 interplay opens a window to refine HCC diagnosis and design new combined therapies.
Keywords: CTR1; MYC; copper; hepatocellular carcinoma; non alcoholic fatty liver disease.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that they have no conflicts of interest.
Figures










References
-
- Hordyjewska A, Ł Popiołek, Kocot J. ‘The many “‘faces’” of copper in medicine and treatment. BioMetals. 2014;27:611–621. https://doi.org/10.1007/s10534-014-9736-5. - DOI - PMC - PubMed
-
- Lowe J, Taveira-da-Silva R, Hilário-Souza E. Dissecting copper homeostasis in diabetes mellitus. IUBMB Life. 2017;69:255–262. https://doi.org/10.1002/iub.1614. - DOI - PubMed
-
- Morrell A, Tallino S, Yu L, Burkhead JL. The role of insufficient copper in lipid synthesis and fatty-liver disease. IUBMB Life. 2017;69:263–270. https://doi.org/10.1002/iub.1613. - DOI - PMC - PubMed
-
- Wachsmann J, Peng F. Molecular imaging and therapy targeting copper metabolism in hepatocellular carcinoma. World Journal of Gastroenterology. 2016;22:221–31. https://doi.org/10.3748/wjg.v22.i1.221. - DOI - PMC - PubMed
-
- Wee NK, Weinstein DC, Fraser ST, Assinder SJ. The mammalian copper transporters CTR1 and CTR2 and their roles in development and disease. Int J Biochem Cell Biol. 2013;45:960–3. https://doi.org/10.1016/j.biocel.2013.01.018. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

