Allele-specific long-distance regulation dictates IL-32 isoform switching and mediates susceptibility to HIV-1
- PMID: 29507875
- PMCID: PMC5833994
- DOI: 10.1126/sciadv.1701729
Allele-specific long-distance regulation dictates IL-32 isoform switching and mediates susceptibility to HIV-1
Abstract
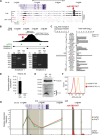
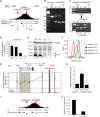
We integrated data obtained from HIV-1 genome-wide association studies with T cell-derived epigenome data and found that the noncoding intergenic variant rs4349147, which is statistically associated with HIV-1 acquisition, is located in a CD4+ T cell-specific deoxyribonuclease I hypersensitive region, suggesting regulatory potential for this variant. Deletion of the rs4349147 element in Jurkat cells strongly reduced expression of interleukin-32 (IL-32), approximately 10-kb upstream, and chromosome conformation capture assays identified a chromatin loop between rs4349147 and the IL-32 promoter validating its function as a long-distance enhancer. We generated single rs4349147-A or rs4349147-G allele clones and demonstrated that IL-32 enhancer activity and interaction with the IL-32 promoter are strongly allele dependent; rs4349147 -/A cells display reduced IL-32 expression and altered chromatin conformation as compared to rs4349147 G/- cells. Moreover, RNA sequencing demonstrated that rs4349147 G/- cells express a lower relative ratio of IL-32α to non-α isoforms than rs4349147 -/A cells and display increased expression of lymphocyte activation factors rendering them more prone to infection with HIV-1. In agreement, in primary CD4+ T cells, both treatment with recombinant IL-32γ (rIL-32γ) but not rIL-32α, and exogenous lentiviral overexpression of IL-32γ or IL-32β but not IL-32α resulted in a proinflammatory T cell cytokine environment concomitant with increased susceptibility to HIV infection. Our data demonstrate that rs4349147-G promotes transcription of non-IL-32α isoforms, generating a proinflammatory environment more conducive to HIV infection. This study provides a mechanistic link between a HIV-associated noncoding DNA variant and the expression of different IL-32 isoforms that display discrete anti-HIV properties.
Figures


Similar articles
-
Upregulation of IL-32 Isoforms in Virologically Suppressed HIV-Infected Individuals: Potential Role in Persistent Inflammation and Transcription From Stable HIV-1 Reservoirs.J Acquir Immune Defic Syndr. 2019 Dec 15;82(5):503-513. doi: 10.1097/QAI.0000000000002185. J Acquir Immune Defic Syndr. 2019. PMID: 31714430 Free PMC article.
-
Inflammation-dependent secretion and splicing of IL-32{gamma} in rheumatoid arthritis.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4962-7. doi: 10.1073/pnas.1016005108. Epub 2011 Mar 7. Proc Natl Acad Sci U S A. 2011. PMID: 21383200 Free PMC article.
-
Alternatively spliced isoforms of IL-32 differentially influence cell death pathways in cancer cell lines.Carcinogenesis. 2016 Feb;37(2):197-205. doi: 10.1093/carcin/bgv172. Epub 2015 Dec 17. Carcinogenesis. 2016. PMID: 26678222
-
Insights into the role of IL-32 in cancer.Semin Immunol. 2018 Aug;38:24-32. doi: 10.1016/j.smim.2018.03.004. Epub 2018 May 8. Semin Immunol. 2018. PMID: 29747940 Review.
-
IL-26, a Cytokine With Roles in Extracellular DNA-Induced Inflammation and Microbial Defense.Front Immunol. 2019 Feb 12;10:204. doi: 10.3389/fimmu.2019.00204. eCollection 2019. Front Immunol. 2019. PMID: 30809226 Free PMC article. Review.
Cited by
-
Targeting TIGIT Inhibits Bladder Cancer Metastasis Through Suppressing IL-32.Front Pharmacol. 2022 Jan 5;12:801493. doi: 10.3389/fphar.2021.801493. eCollection 2021. Front Pharmacol. 2022. PMID: 35069212 Free PMC article.
-
A Critical Overview of Interleukin 32 in Leishmaniases.Front Immunol. 2022 Mar 3;13:849340. doi: 10.3389/fimmu.2022.849340. eCollection 2022. Front Immunol. 2022. PMID: 35309341 Free PMC article. Review.
-
A Two-Color Haploid Genetic Screen Identifies Novel Host Factors Involved in HIV-1 Latency.mBio. 2021 Dec 21;12(6):e0298021. doi: 10.1128/mBio.02980-21. Epub 2021 Dec 7. mBio. 2021. PMID: 34872356 Free PMC article.
-
Upregulation of IL-32 Isoforms in Virologically Suppressed HIV-Infected Individuals: Potential Role in Persistent Inflammation and Transcription From Stable HIV-1 Reservoirs.J Acquir Immune Defic Syndr. 2019 Dec 15;82(5):503-513. doi: 10.1097/QAI.0000000000002185. J Acquir Immune Defic Syndr. 2019. PMID: 31714430 Free PMC article.
-
The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases.BMC Med Genomics. 2022 Apr 1;15(1):74. doi: 10.1186/s12920-022-01216-w. BMC Med Genomics. 2022. PMID: 35365203 Free PMC article.
References
-
- Wang X., Tucker N. R., Rizki G., Mills R., Krijger P. H. L., de Wit E., Subramanian V., Bartell E., Nguyen X.-X., Ye J., Leyton-Mange J., Dolmatova E. V., van der Harst P., de Laat W., Ellinor P. T., Newton-Cheh C., Milan D. J., Kellis M., Boyer L. A., Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. eLife 5, e10557 (2016). - PMC - PubMed
-
- Lingappa J. R., Petrovski S., Kahle E., Fellay J., Shianna K., McElrath M. J., Thomas K. K., Baeten J. M., Celum C., Wald A., de Bruyn G., Mullins J. I., Nakku-Joloba E., Farquhar C., Essex M., Donnell D., Kiarie J., Haynes B., Goldstein D. the Partners in Prevention HSV/HIV Transmission Study Team , Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLOS ONE 6, e28632 (2011). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

