CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres
- PMID: 29509805
- PMCID: PMC5839545
- DOI: 10.1371/journal.pone.0192572
CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres
Abstract
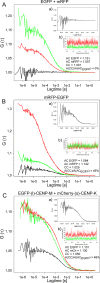
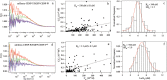
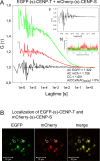
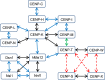
Kinetochore proteins assemble onto centromeric chromatin and regulate DNA segregation during cell division. The inner kinetochore proteins bind centromeres while most outer kinetochore proteins assemble at centromeres during mitosis, connecting the complex to microtubules. Here, we measured the co-migration between protein pairs of the constitutive centromere associated network (CCAN) and hMis12 complexes by fluorescence cross-correlation spectroscopy (FCCS) in the nucleoplasm outside centromeres in living human interphase cells. FCCS is a method that can tell if in living cells two differently fluorescently labelled molecules migrate independently, or co-migrate and thus are part of one and the same soluble complex. We also determined the apparent dissociation constants (Kd) of the hetero-dimers CENP-T/W and CENP-S/X. We measured co-migration between CENP-K and CENP-T as well as between CENP-M and CENP-T but not between CENP-T/W and CENP-S/X. Furthermore, CENP-C co-migrated with CENP-H, and CENP-K with CENP-N as well as with CENP-L. Thus, in the nucleoplasm outside centromeres, a large fraction of the CENP-H/I/K/M proteins interact with CENP-C, CENP-N/L and CENP-T/W but not with CENP-S/X. Our FCCS analysis of the Mis12 complex showed that hMis12, Nsl1, Dsn1 and Nnf1 also form a complex outside centromeres of which at least hMis12 associated with the CENP-C/H/I/K/M/T/W/N/L complex.
Conflict of interest statement
Figures

References
-
- Pesenti M, Weir JR, Musacchio A (2016) Progress in the structural and functional characterization of kinetochores. Curr Opin Struc Biol 37:152–163. - PubMed
-
- Nagpal H, Fukagawa T (2016) Kinetochore assembly and function through the cell cycle. Chromosoma 125:645–659. doi: 10.1007/s00412-016-0608-3 - DOI - PubMed
-
- McKinley KL, Cheeseman I.M (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17:16–29. doi: 10.1038/nrm.2015.5 - DOI - PMC - PubMed
-
- Schalch T, Steiner FA (2016) Structure of centromere chromatin: from nucleosome to chromosomal architecture. Chromosoma, doi: 10.1007/s00412-016-0620-7 - DOI - PMC - PubMed
-
- Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28:2511–2531. doi: 10.1038/emboj.2009.173 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

