Dysregulation of TBX1 dosage in the anterior heart field results in congenital heart disease resembling the 22q11.2 duplication syndrome
- PMID: 29509905
- PMCID: PMC5961083
- DOI: 10.1093/hmg/ddy078
Dysregulation of TBX1 dosage in the anterior heart field results in congenital heart disease resembling the 22q11.2 duplication syndrome
Abstract
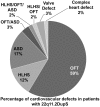
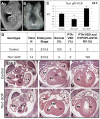
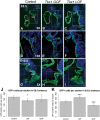
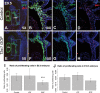
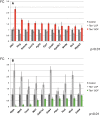
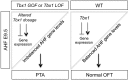
Non-allelic homologous recombination events on chromosome 22q11.2 during meiosis can result in either the deletion (22q11.2DS) or duplication (22q11.2DupS) syndrome. Although the spectrum and frequency of congenital heart disease (CHD) are known for 22q11.2DS, there is less known for 22q11.2DupS. We now evaluated cardiac phenotypes in 235 subjects with 22q11.2DupS including 102 subjects we collected and 133 subjects that were previously reported as a confirmation and found 25% have CHD, mostly affecting the cardiac outflow tract (OFT). Previous studies have shown that global loss or gain of function (LOF; GOF) of mouse Tbx1, encoding a T-box transcription factor mapping to the region of synteny to 22q11.2, results in similar OFT defects. To further evaluate Tbx1 function in the progenitor cells forming the cardiac OFT, termed the anterior heart field, Tbx1 was overexpressed using the Mef2c-AHF-Cre driver (Tbx1 GOF). Here we found that all resulting conditional GOF embryos had a persistent truncus arteriosus (PTA), similar to what was previously reported for conditional Tbx1 LOF mutant embryos. To understand the basis for the PTA in the conditional GOF embryos, we found that proliferation in the Mef2c-AHF-Cre lineage cells before migrating to the heart, was reduced and critical genes were oppositely changed in this tissue in Tbx1 GOF embryos versus conditional LOF embryos. These results suggest that a major function of TBX1 in the AHF is to maintain the normal balance of expression of key cardiac developmental genes required to form the aorta and pulmonary trunk, which is disrupted in 22q11.2DS and 22q11.2DupS.
Figures
Similar articles
-
Reduced dosage of β-catenin provides significant rescue of cardiac outflow tract anomalies in a Tbx1 conditional null mouse model of 22q11.2 deletion syndrome.PLoS Genet. 2017 Mar 27;13(3):e1006687. doi: 10.1371/journal.pgen.1006687. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28346476 Free PMC article.
-
Left pulmonary artery in 22q11.2 deletion syndrome. Echocardiographic evaluation in patients without cardiac defects and role of Tbx1 in mice.PLoS One. 2019 Apr 1;14(4):e0211170. doi: 10.1371/journal.pone.0211170. eCollection 2019. PLoS One. 2019. PMID: 30933971 Free PMC article.
-
A loss-of-function mutation p.T52S in RIPPLY3 is a potential predisposing genetic risk factor for Chinese Han conotruncal heart defect patients without the 22q11.2 deletion/duplication.J Transl Med. 2018 Sep 21;16(1):260. doi: 10.1186/s12967-018-1633-1. J Transl Med. 2018. PMID: 30241482 Free PMC article.
-
Understanding the role of Tbx1 as a candidate gene for 22q11.2 deletion syndrome.Curr Allergy Asthma Rep. 2013 Dec;13(6):613-21. doi: 10.1007/s11882-013-0384-6. Curr Allergy Asthma Rep. 2013. PMID: 23996541 Free PMC article. Review.
-
22q11 deletion syndrome: a role for TBX1 in pharyngeal and cardiovascular development.Pediatr Cardiol. 2010 Apr;31(3):378-90. doi: 10.1007/s00246-009-9613-0. Pediatr Cardiol. 2010. PMID: 20054531 Review.
Cited by
-
The Transitional Heart: From Early Embryonic and Fetal Development to Neonatal Life.Fetal Diagn Ther. 2020;47(5):373-386. doi: 10.1159/000501906. Epub 2019 Sep 18. Fetal Diagn Ther. 2020. PMID: 31533099 Free PMC article. Review.
-
Genetic Basis of Human Congenital Heart Disease.Cold Spring Harb Perspect Biol. 2020 Sep 1;12(9):a036749. doi: 10.1101/cshperspect.a036749. Cold Spring Harb Perspect Biol. 2020. PMID: 31818857 Free PMC article.
-
Mending a broken heart: In vitro, in vivo and in silico models of congenital heart disease.Dis Model Mech. 2021 Mar 28;14(3):dmm047522. doi: 10.1242/dmm.047522. Dis Model Mech. 2021. PMID: 33787508 Free PMC article. Review.
-
HNRNPR Variants that Impair Homeobox Gene Expression Drive Developmental Disorders in Humans.Am J Hum Genet. 2019 Jun 6;104(6):1040-1059. doi: 10.1016/j.ajhg.2019.03.024. Epub 2019 May 9. Am J Hum Genet. 2019. PMID: 31079900 Free PMC article.
-
New Concepts in the Development and Malformation of the Arterial Valves.J Cardiovasc Dev Dis. 2020 Sep 24;7(4):38. doi: 10.3390/jcdd7040038. J Cardiovasc Dev Dis. 2020. PMID: 32987700 Free PMC article. Review.
References
-
- Gross S.J., Stosic M., McDonald-McGinn D.M., Bassett A.S., Norvez A., Dhamankar R., Kobara K., Kirkizlar E., Zimmermann B., Wayham N.. et al. (2016) Clinical experience with single-nucleotide polymorphism-based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet Gynecol., 47, 177–183. - PMC - PubMed
-
- Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E. (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet., 8, 1157–1167. - PubMed
-
- Shaikh T.H., Kurahashi H., Emanuel B.S. (2001) Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: an update and literature review. Genet. Med., 3, 6–13. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

