Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana
- PMID: 29509983
- PMCID: PMC6131417
- DOI: 10.1111/pbi.12916
Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana
Abstract
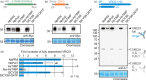
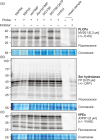
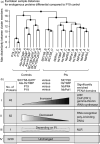
Agroinfiltrated Nicotiana benthamiana is a flexible and scalable platform for recombinant protein (RP) production, but its great potential is hampered by plant proteases that degrade RPs. Here, we tested 29 candidate protease inhibitors (PIs) in agroinfiltrated N. benthamiana leaves for enhancing accumulation of three unrelated RPs: glycoenzyme α-Galactosidase; glycohormone erythropoietin (EPO); and IgG antibody VRC01. Of the previously described PIs enhancing RP accumulation, we found only cystatin SlCYS8 to be effective. We identified three additional new, unrelated PIs that enhance RP accumulation: N. benthamiana NbPR4, NbPot1 and human HsTIMP, which have been reported to inhibit cysteine, serine and metalloproteases, respectively. Remarkably, accumulation of all three RPs is enhanced by each PI similarly, suggesting that the mechanism of degradation of unrelated RPs follows a common pathway. Inhibitory functions HsTIMP and SlCYS8 are required to enhance RP accumulation, suggesting that their target proteases may degrade RPs. Different PIs additively enhance RP accumulation, but the effect of each PI is dose-dependent. Activity-based protein profiling (ABPP) revealed that the activities of papain-like Cys proteases (PLCPs), Ser hydrolases (SHs) or vacuolar processing enzymes (VPEs) in leaves are unaffected upon expression of the new PIs, whereas SlCYS8 expression specifically suppresses PLCP activity only. Quantitative proteomics indicates that the three new PIs affect agroinfiltrated tissues similarly and that they all increase immune responses. NbPR4, NbPot1 and HsTIMP can be used to study plant proteases and improve RP accumulation in molecular farming.
Keywords: Agrobacterium tumefaciens; Nicotiana benthamiana; activity-based protein profiling; agroinfiltration; protease inhibitor; transient expression.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures




References
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Bombarely, A. , Rosli, H.G. , Vrebalov, J. , Moffett, P. , Mueller, L.A. and Martin, G.B. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant‐microbe biology research. Mol. Plant Microbe Interact. 25, 1523–1530. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

