Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality
- PMID: 29509985
- PMCID: PMC6131414
- DOI: 10.1111/pbi.12910
Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality
Abstract
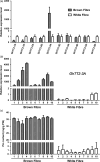
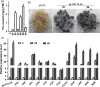
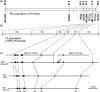
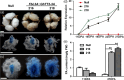
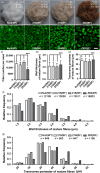
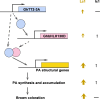
Brown cotton fibres are the most widely used naturally coloured raw materials for the eco-friendly textile industry. Previous studies have indicated that brown fibre pigments belong to proanthocyanidins (PAs) or their derivatives, and fibre coloration is negatively associated with cotton productivity and fibre quality. To date, the molecular basis controlling the biosynthesis and accumulation of brown pigments in cotton fibres is largely unknown. In this study, based on expressional and transgenic analyses of cotton homologs of ArabidopsisPA regulator TRANSPARENT TESTA 2 (TT2) and fine-mapping of the cotton dark-brown fibre gene (Lc1), we show that a TT2 homolog, GhTT2-3A, controls PA biosynthesis and brown pigmentation in cotton fibres. We observed that GhTT2-3A activated GhbHLH130D, a homolog of ArabidopsisTT8, which in turn synergistically acted with GhTT2-3A to activate downstream PA structural genes and PA synthesis and accumulation in cotton fibres. Furthermore, the up-regulation of GhTT2-3A in fibres at the secondary wall-thickening stage resulted in brown mature fibres, and fibre quality and lint percentage were comparable to that of the white-fibre control. The findings of this study reveal the regulatory mechanism controlling brown pigmentation in cotton fibres and demonstrate a promising biotechnological strategy to break the negative linkage between coloration and fibre quality and/or productivity.
Keywords: Gossypium; TT2; brown fibre; proanthocyanidin; transgenic cotton.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

References
-
- Appelhagen, I. , Thiedig, K. , Nordholt, N. , Schmidt, N. , Huep, G. , Sagasser, M. and Weisshaar, B. (2014) Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta, 240, 955–970. - PubMed
-
- Appelhagen, I. , Nordholt, N. , Seidel, T. , Spelt, K. , Koes, R. , Quattrochio, F. , Sagasser, M. et al. (2015) TRANSPARENT TESTA 13 is a tonoplast P3A ‐ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82, 840–849. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 30971713 /National Natural Science Foundation of China/International
- 2016ZX08005005-001/Genetically Modified Organisms Breeding Major Project of China/International
- 2016E02001/Special Project of Regional Collaborative Innovation in the Xinjiang Uygur Autonomous Region/International
- 31571582 /National Natural Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

