Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group
- PMID: 29509989
- PMCID: PMC5947300
- DOI: 10.1111/mec.14546
Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group
Abstract
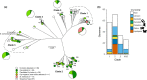
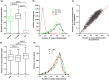
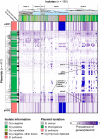
Bacterial plasmids can vary from small selfish genetic elements to large autonomous replicons that constitute a significant proportion of total cellular DNA. By conferring novel function to the cell, plasmids may facilitate evolution but their mobility may be opposed by co-evolutionary relationships with chromosomes or encouraged via the infectious sharing of genes encoding public goods. Here, we explore these hypotheses through large-scale examination of the association between plasmids and chromosomal DNA in the phenotypically diverse Bacillus cereus group. This complex group is rich in plasmids, many of which encode essential virulence factors (Cry toxins) that are known public goods. We characterized population genomic structure, gene content and plasmid distribution to investigate the role of mobile elements in diversification. We analysed coding sequence within the core and accessory genome of 190 B. cereus group isolates, including 23 novel sequences and genes from 410 reference plasmid genomes. While cry genes were widely distributed, those with invertebrate toxicity were predominantly associated with one sequence cluster (clade 2) and phenotypically defined Bacillus thuringiensis. Cry toxin plasmids in clade 2 showed evidence of recent horizontal transfer and variable gene content, a pattern of plasmid segregation consistent with transfer during infectious cooperation. Nevertheless, comparison between clades suggests that co-evolutionary interactions may drive association between plasmids and chromosomes and limit wider transfer of key virulence traits. Proliferation of successful plasmid and chromosome combinations is a feature of specialized pathogens with characteristic niches (Bacillus anthracis, B. thuringiensis) and has occurred multiple times in the B. cereus group.
Keywords: Bacillus cereus; Bacillus thuringiensis; insecticidal toxins; mobile genetic elements; pan-genome.
© 2018 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures

References
-
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 - DOI - PubMed
-
- Antonation, K. S. , Grutzmacher, K. , Dupke, S. , Mabon, P. , Zimmermann, F. , Lankester, F. , … de Nys, H. M. (2016). Bacillus cereus biovar anthracis causing anthrax in sub‐saharan Africa‐chromosomal monophyly and broad geographic distribution. PLoS Neglected Tropical Diseases, 10, e0004923. - PMC - PubMed
-
- Aziz, R. K. , Bartels, D. , Best, A. A. , DeJongh, M. , Disz, T. , Edwards, R. A. , … Meyer, F. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics, 9, 75 https://doi.org/10.1186/1471-2164-9-75 - DOI - PMC - PubMed
-
- Berry, C. , O'Neil, S. , Ben‐Dov, E. , Jones, A. F. , Murphy, L. , Quail, M. A. , … Parkhill, J. (2002). Complete sequence and organization of pBtoxis the toxin‐coding plasmid of Bacillus thuringiensis subsp. israelensis . Applied and Environment Microbiology, 68, 5082–5095. https://doi.org/10.1128/AEM.68.10.5082-5095.2002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

