Discovery of the Tyrobetaine Natural Products and Their Biosynthetic Gene Cluster via Metabologenomics
- PMID: 29510029
- PMCID: PMC5944846
- DOI: 10.1021/acschembio.7b01089
Discovery of the Tyrobetaine Natural Products and Their Biosynthetic Gene Cluster via Metabologenomics
Abstract
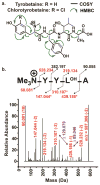
Natural products (NPs) are a rich source of medicines, but traditional discovery methods are often unsuccessful due to high rates of rediscovery. Genetic approaches for NP discovery are promising, but progress has been slow due to the difficulty of identifying unique biosynthetic gene clusters (BGCs) and poor gene expression. We previously developed the metabologenomics method, which combines genomic and metabolomic data to discover new NPs and their BGCs. Here, we utilize metabologenomics in combination with molecular networking to discover a novel class of NPs, the tyrobetaines: nonribosomal peptides with an unusual trimethylammonium tyrosine residue. The BGC for this unusual class of compounds was identified using metabologenomics and computational structure prediction data. Heterologous expression confirmed the BGC and suggests an unusual mechanism for trimethylammonium formation. Overall, the discovery of the tyrobetaines shows the great potential of metabologenomics combined with molecular networking and computational structure prediction for identifying interesting biosynthetic reactions and novel NPs.
Conflict of interest statement
The authors declare the following competing financial interest(s): Anthony W. Goering, Regan J. Thomson, Neil L. Kelleher, and William W. Metcalf have a financial interest in metablogenomics via Microbial Pharmaceuticals. These potential conflicts of interests are managed through the respective university’s conflict of interest policy.
Figures

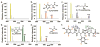

References
-
- Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. - PubMed
-
- Cragg GM, Grothaus PG, Newman DJ. Impact of Natural Products on Developing New Anti-Cancer Agents. Chem Rev. 2009;109:3012–3043. - PubMed
-
- Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discovery. 2015;14:111–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

