Loss of stomach, loss of appetite? Sequencing of the ballan wrasse (Labrus bergylta) genome and intestinal transcriptomic profiling illuminate the evolution of loss of stomach function in fish
- PMID: 29510660
- PMCID: PMC5840709
- DOI: 10.1186/s12864-018-4570-8
Loss of stomach, loss of appetite? Sequencing of the ballan wrasse (Labrus bergylta) genome and intestinal transcriptomic profiling illuminate the evolution of loss of stomach function in fish
Abstract
Background: The ballan wrasse (Labrus bergylta) belongs to a large teleost family containing more than 600 species showing several unique evolutionary traits such as lack of stomach and hermaphroditism. Agastric fish are found throughout the teleost phylogeny, in quite diverse and unrelated lineages, indicating stomach loss has occurred independently multiple times in the course of evolution. By assembling the ballan wrasse genome and transcriptome we aimed to determine the genetic basis for its digestive system function and appetite regulation. Among other, this knowledge will aid the formulation of aquaculture diets that meet the nutritional needs of agastric species.
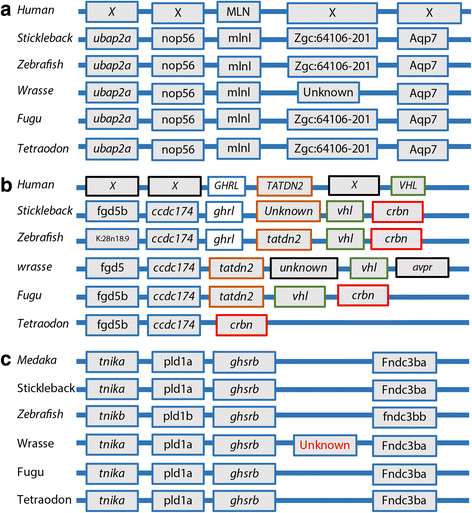
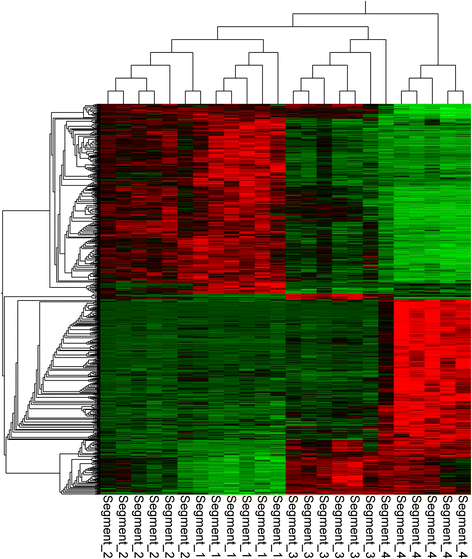
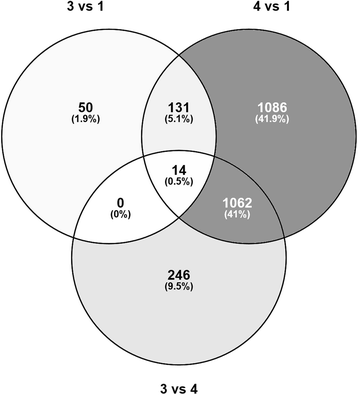
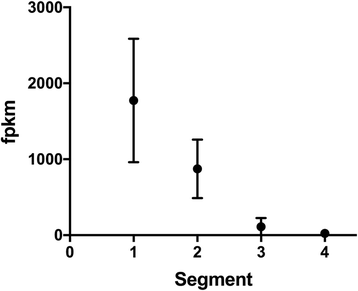
Results: Long and short read sequencing technologies were combined to generate a ballan wrasse genome of 805 Mbp. Analysis of the genome and transcriptome assemblies confirmed the absence of genes that code for proteins involved in gastric function. The gene coding for the appetite stimulating protein ghrelin was also absent in wrasse. Gene synteny mapping identified several appetite-controlling genes and their paralogs previously undescribed in fish. Transcriptome profiling along the length of the intestine found a declining expression gradient from the anterior to the posterior, and a distinct expression profile in the hind gut.
Conclusions: We showed gene loss has occurred for all known genes related to stomach function in the ballan wrasse, while the remaining functions of the digestive tract appear intact. The results also show appetite control in ballan wrasse has undergone substantial changes. The loss of ghrelin suggests that other genes, such as motilin, may play a ghrelin like role. The wrasse genome offers novel insight in to the evolutionary traits of this large family. As the stomach plays a major role in protein digestion, the lack of genes related to stomach digestion in wrasse suggests it requires formulated diets with higher levels of readily digestible protein than those for gastric species.
Keywords: Appetite; Ballan wrasse; Digestion; Ghrelin; Intestine; Labrid; Motilin; Neuropeptide; Stomach; Transcobalamin.
Conflict of interest statement
Ethics approval and consent to participate
The fish were collected from a commercial hatchery (Marine Harvest Labrus, Rong, Norway), where ballan wrasse are reared in accordance with the Norwegian Animal Welfare Act of 12 December 1974, no. 73, §§22 and 30, amended 19 June 2009. The facility has a general permission to rear all developmental stages of Labrus berggylta, license number H ØN0038 provided by the Norwegian Directorate of fisheries (
Since fish were sampled from the production line and did not undergo any treatment or handling except for sampling a special approval from the food authorities and ethics committee is deemed unnecessary according to Norwegian regulations “Legislation on the use of animals” § 6 “Approval of study” (English translation)(
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Dietary Lipid Modulation of Intestinal Serotonin in Ballan Wrasse (Labrus bergylta)-In Vitro Analyses.Front Endocrinol (Lausanne). 2021 Mar 23;12:560055. doi: 10.3389/fendo.2021.560055. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33833735 Free PMC article.
-
Effects of Cholecystokinin (CCK) on Gut Motility in the Stomachless Fish Ballan Wrasse (Labrus bergylta).Front Neurosci. 2019 Jun 7;13:553. doi: 10.3389/fnins.2019.00553. eCollection 2019. Front Neurosci. 2019. PMID: 31231179 Free PMC article.
-
Digestive and immune functions in the intestine of wild Ballan wrasse (Labrus bergylta).Comp Biochem Physiol A Mol Integr Physiol. 2021 Oct;260:111011. doi: 10.1016/j.cbpa.2021.111011. Epub 2021 Jun 24. Comp Biochem Physiol A Mol Integr Physiol. 2021. PMID: 34174428
-
Sustainable production and use of cleaner fish for the biological control of sea lice: recent advances and current challenges.Vet Rec. 2018 Sep 29;183(12):383. doi: 10.1136/vr.104966. Epub 2018 Jul 30. Vet Rec. 2018. PMID: 30061113 Review.
-
A review of potential pathogens of sea lice and the application of cleaner fish in biological control.Pest Manag Sci. 2002 Jun;58(6):546-58. doi: 10.1002/ps.509. Pest Manag Sci. 2002. PMID: 12138621 Review.
Cited by
-
Dietary Lipid Modulation of Intestinal Serotonin in Ballan Wrasse (Labrus bergylta)-In Vitro Analyses.Front Endocrinol (Lausanne). 2021 Mar 23;12:560055. doi: 10.3389/fendo.2021.560055. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33833735 Free PMC article.
-
Soya saponins and prebiotics alter intestinal functions in Ballan wrasse (Labrus bergylta).Br J Nutr. 2023 Sep 14;130(5):765-782. doi: 10.1017/S000711452200383X. Epub 2023 Jan 12. Br J Nutr. 2023. PMID: 36632013 Free PMC article.
-
Diverse Origins of Near-Identical Antifreeze Proteins in Unrelated Fish Lineages Provide Insights Into Evolutionary Mechanisms of New Gene Birth and Protein Sequence Convergence.Mol Biol Evol. 2024 Sep 4;41(9):msae182. doi: 10.1093/molbev/msae182. Mol Biol Evol. 2024. PMID: 39213383 Free PMC article.
-
Gut Immune System and the Implications of Oral-Administered Immunoprophylaxis in Finfish Aquaculture.Front Immunol. 2021 Dec 16;12:773193. doi: 10.3389/fimmu.2021.773193. eCollection 2021. Front Immunol. 2021. PMID: 34975860 Free PMC article. Review.
-
Effects of Cholecystokinin (CCK) on Gut Motility in the Stomachless Fish Ballan Wrasse (Labrus bergylta).Front Neurosci. 2019 Jun 7;13:553. doi: 10.3389/fnins.2019.00553. eCollection 2019. Front Neurosci. 2019. PMID: 31231179 Free PMC article.
References
-
- Parenti P, Randall JE. Bioline international official site (site up-dated regularly) Ichthyol Bull. 2000;68:1–97.
-
- Artüz ML: Age and growth of the ballan wrasse Labrus bergylta Ascanius 1767. 2007:1–5.
-
- Bjordal A: Sea lice infestation on farmed salmon: possible use of cleaner-fish as an alternative method for de-lousing. Canadian technical report of fisheries and aquatic … 1990.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

