Characterization of the human RFX transcription factor family by regulatory and target gene analysis
- PMID: 29510665
- PMCID: PMC5838959
- DOI: 10.1186/s12864-018-4564-6
Characterization of the human RFX transcription factor family by regulatory and target gene analysis
Abstract
Background: Evolutionarily conserved RFX transcription factors (TFs) regulate their target genes through a DNA sequence motif called the X-box. Thereby they regulate cellular specialization and terminal differentiation. Here, we provide a comprehensive analysis of all the eight human RFX genes (RFX1-8), their spatial and temporal expression profiles, potential upstream regulators and target genes.
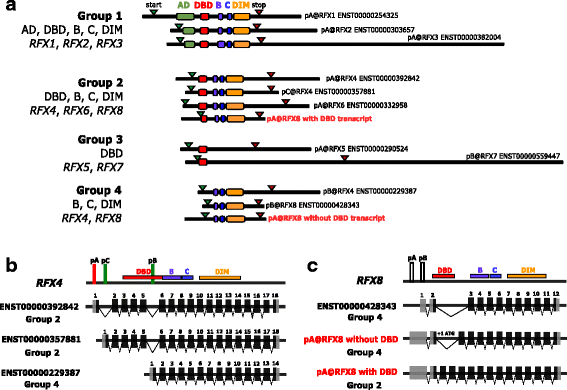
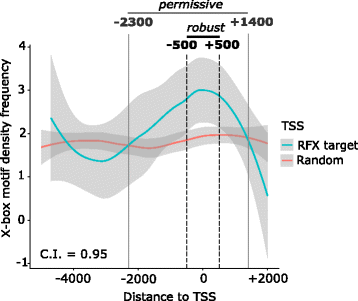
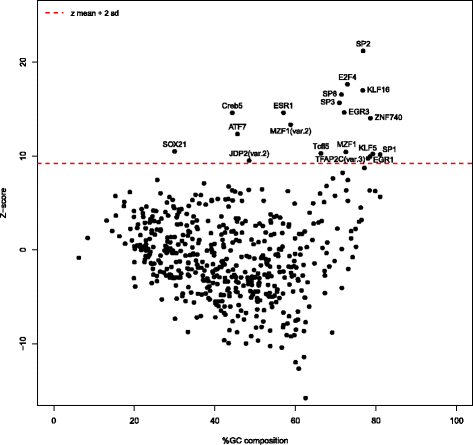
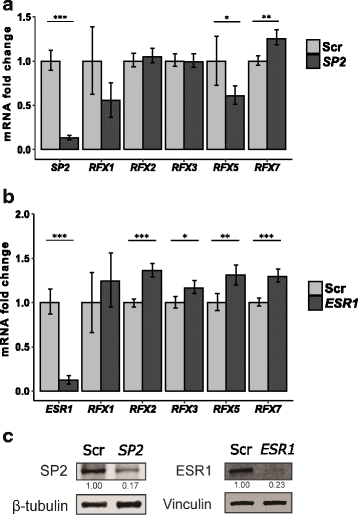
Results: We extracted all known human RFX1-8 gene expression profiles from the FANTOM5 database derived from transcription start site (TSS) activity as captured by Cap Analysis of Gene Expression (CAGE) technology. RFX genes are broadly (RFX1-3, RFX5, RFX7) and specifically (RFX4, RFX6) expressed in different cell types, with high expression in four organ systems: immune system, gastrointestinal tract, reproductive system and nervous system. Tissue type specific expression profiles link defined RFX family members with the target gene batteries they regulate. We experimentally confirmed novel TSS locations and characterized the previously undescribed RFX8 to be lowly expressed. RFX tissue and cell type specificity arises mainly from differences in TSS architecture. RFX transcript isoforms lacking a DNA binding domain (DBD) open up new possibilities for combinatorial target gene regulation. Our results favor a new grouping of the RFX family based on protein domain composition. We uncovered and experimentally confirmed the TFs SP2 and ESR1 as upstream regulators of specific RFX genes. Using TF binding profiles from the JASPAR database, we determined relevant patterns of X-box motif positioning with respect to gene TSS locations of human RFX target genes.
Conclusions: The wealth of data we provide will serve as the basis for precisely determining the roles RFX TFs play in human development and disease.
Keywords: Cell cycle control; Cell differentiation; Cilia; Immune cell proliferation; Neuronal development; Spermatogenesis; Tumor suppression.
Conflict of interest statement
Ethics approval and consent to participate
For work with mammalian cell cultures the authors are in possession of the applicable permits for carrying out studies with genetically modified micro-organisms / GMMs (Dnr 5.8.18–1012/14; Dnr 5.5.18–6998/15).
Consent for publication
This section is not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- KID funding/Karolinska Institutet/International
- Distinguished Professor Award/Karolinska Institutet/International
- Strategic Neurosciences Program/Karolinska Institutet/International
- EU Horizon 2020 Marie Curie Individual Fellowship/Horizon 2020/International
- 174CDE/Genome Canada Large Scale Applied Research Grant/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

