Building protein-protein interaction networks for Leishmania species through protein structural information
- PMID: 29510668
- PMCID: PMC5840830
- DOI: 10.1186/s12859-018-2105-6
Building protein-protein interaction networks for Leishmania species through protein structural information
Abstract
Background: Systematic analysis of a parasite interactome is a key approach to understand different biological processes. It makes possible to elucidate disease mechanisms, to predict protein functions and to select promising targets for drug development. Currently, several approaches for protein interaction prediction for non-model species incorporate only small fractions of the entire proteomes and their interactions. Based on this perspective, this study presents an integration of computational methodologies, protein network predictions and comparative analysis of the protozoan species Leishmania braziliensis and Leishmania infantum. These parasites cause Leishmaniasis, a worldwide distributed and neglected disease, with limited treatment options using currently available drugs.
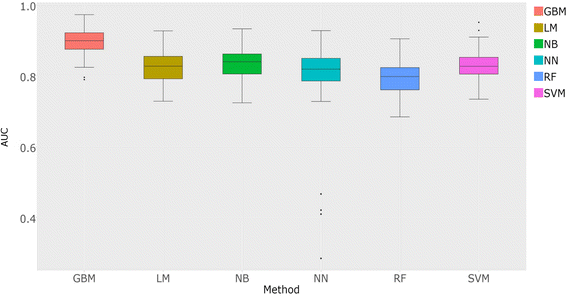
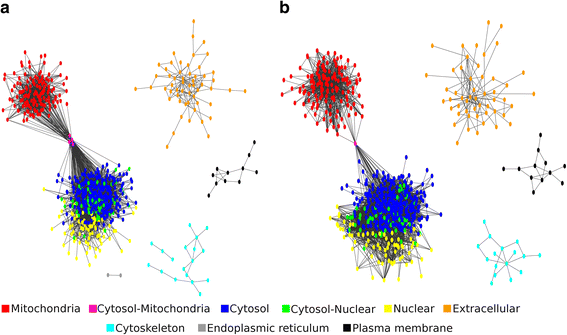
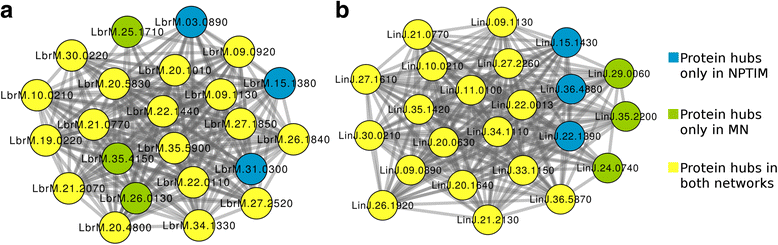
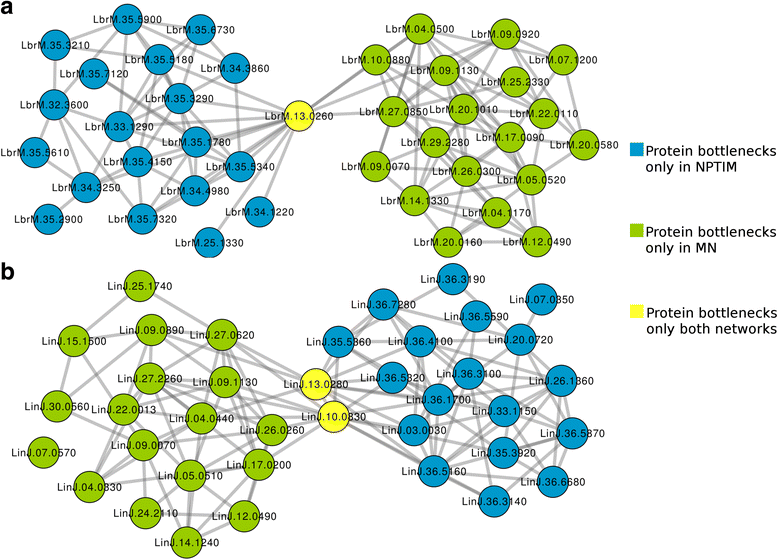
Results: The predicted interactions were obtained from a meta-approach, applying rigid body docking tests and template-based docking on protein structures predicted by different comparative modeling techniques. In addition, we trained a machine-learning algorithm (Gradient Boosting) using docking information performed on a curated set of positive and negative protein interaction data. Our final model obtained an AUC = 0.88, with recall = 0.69, specificity = 0.88 and precision = 0.83. Using this approach, it was possible to confidently predict 681 protein structures and 6198 protein interactions for L. braziliensis, and 708 protein structures and 7391 protein interactions for L. infantum. The predicted networks were integrated to protein interaction data already available, analyzed using several topological features and used to classify proteins as essential for network stability.
Conclusions: The present study allowed to demonstrate the importance of integrating different methodologies of interaction prediction to increase the coverage of the protein interaction of the studied protocols, besides it made available protein structures and interactions not previously reported.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

