Summarizing performance for genome scale measurement of miRNA: reference samples and metrics
- PMID: 29510677
- PMCID: PMC5838960
- DOI: 10.1186/s12864-018-4496-1
Summarizing performance for genome scale measurement of miRNA: reference samples and metrics
Abstract
Background: The potential utility of microRNA as biomarkers for early detection of cancer and other diseases is being investigated with genome-scale profiling of differentially expressed microRNA. Processes for measurement assurance are critical components of genome-scale measurements. Here, we evaluated the utility of a set of total RNA samples, designed with between-sample differences in the relative abundance of miRNAs, as process controls.
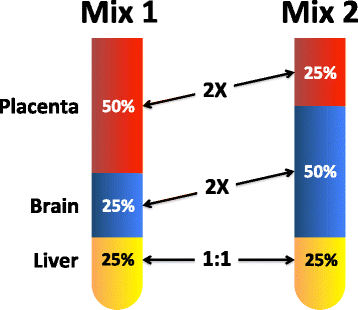
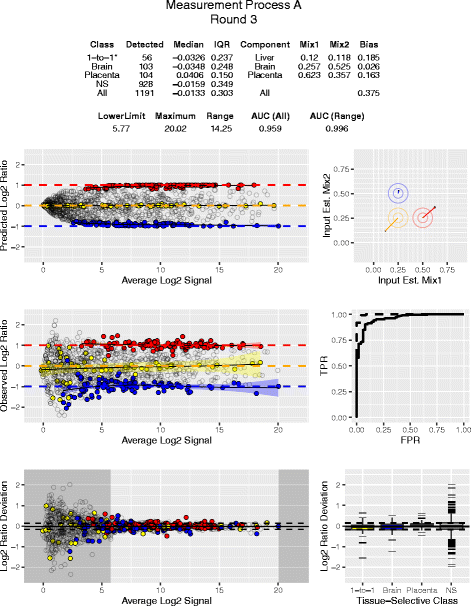
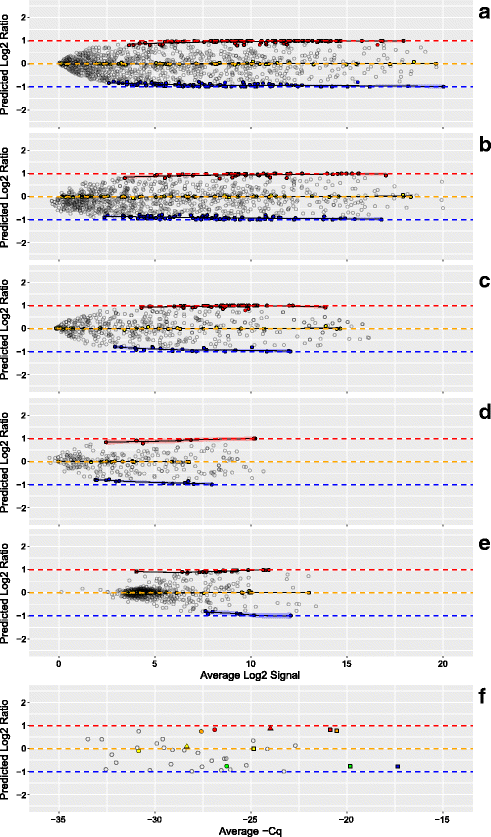
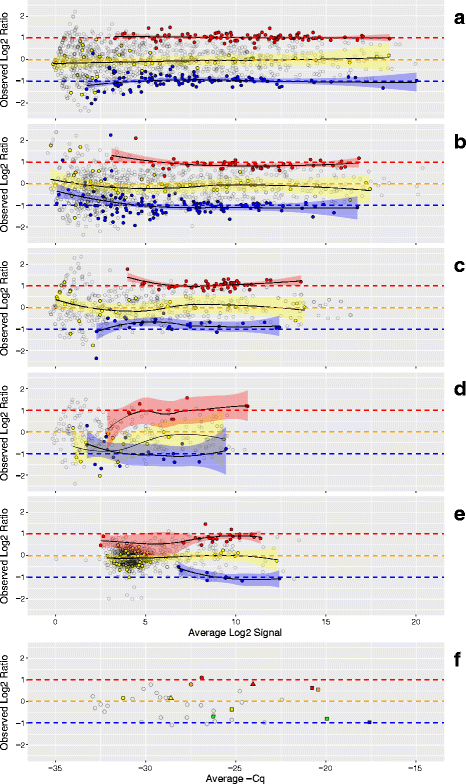
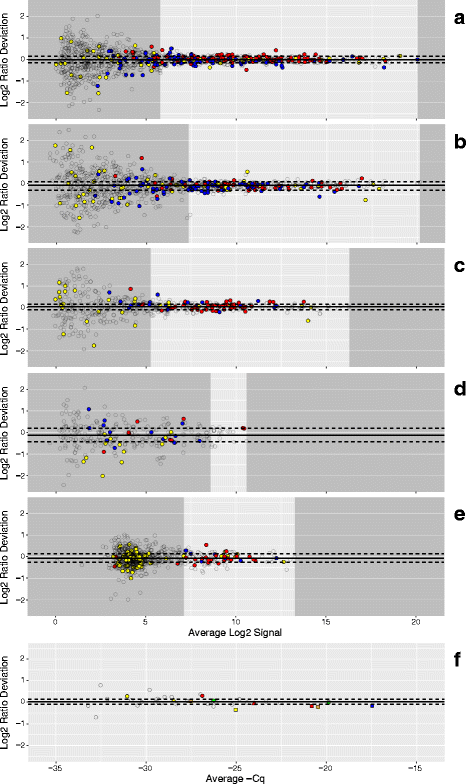
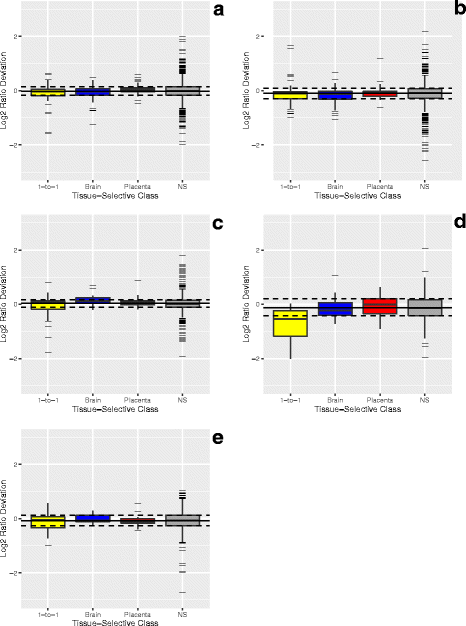
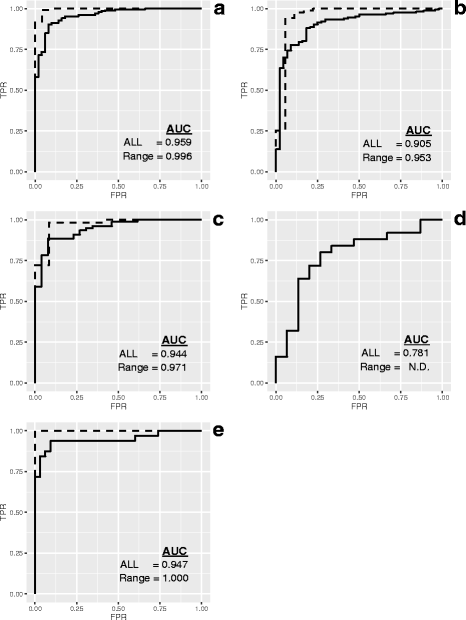
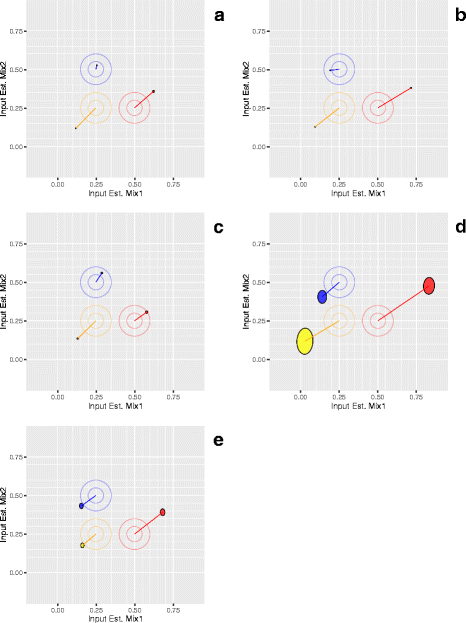
Results: Three pure total human RNA samples (brain, liver, and placenta) and two different mixtures of these components were evaluated as measurement assurance control samples on multiple measurement systems at multiple sites and over multiple rounds. In silico modeling of mixtures provided benchmark values for comparison with physical mixtures. Biomarker development laboratories using next-generation sequencing (NGS) or genome-scale hybridization assays participated in the study and returned data from the samples using their routine workflows. Multiplexed and single assay reverse-transcription PCR (RT-PCR) was used to confirm in silico predicted sample differences. Data visualizations and summary metrics for genome-scale miRNA profiling assessment were developed using this dataset, and a range of performance was observed. These metrics have been incorporated into an online data analysis pipeline and provide a convenient dashboard view of results from experiments following the described design. The website also serves as a repository for the accumulation of performance values providing new participants in the project an opportunity to learn what may be achievable with similar measurement processes.
Conclusions: The set of reference samples used in this study provides benchmark values suitable for assessing genome-scale miRNA profiling processes. Incorporation of these metrics into an online resource allows laboratories to periodically evaluate their performance and assess any changes introduced into their measurement process.
Keywords: Dashboard; Process controls; Reference samples; miRNA; microRNA.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. Human Brain Reference RNA (Cat. No. AM6050), Human Liver Total RNA (Cat. No. AM7960), and Human Placenta Total RNA (Cat. No. AM7950) was obtained from Ambion (Thermo Fisher Scientific).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- U01CA214195/Division of Cancer Prevention, National Cancer Institute/International
- T32 HL007035/HL/NHLBI NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- U24CA115091/Division of Cancer Prevention, National Cancer Institute/International
- U01CA214182/Division of Cancer Prevention, National Cancer Institute/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

