A missense mutation in MYH1 is associated with susceptibility to immune-mediated myositis in Quarter Horses
- PMID: 29510741
- PMCID: PMC5838957
- DOI: 10.1186/s13395-018-0155-0
A missense mutation in MYH1 is associated with susceptibility to immune-mediated myositis in Quarter Horses
Abstract
Background: The cause of immune-mediated myositis (IMM), characterized by recurrent, rapid-onset muscle atrophy in Quarter Horses (QH), is unknown. The histopathologic hallmark of IMM is lymphocytic infiltration of myofibers. The purpose of this study was to identify putative functional variants associated with equine IMM.
Methods: A genome-wide association (GWA) study was performed on 36 IMM QHs and 54 breed matched unaffected QHs from the same environment using the Equine SNP50 and SNP70 genotyping arrays.
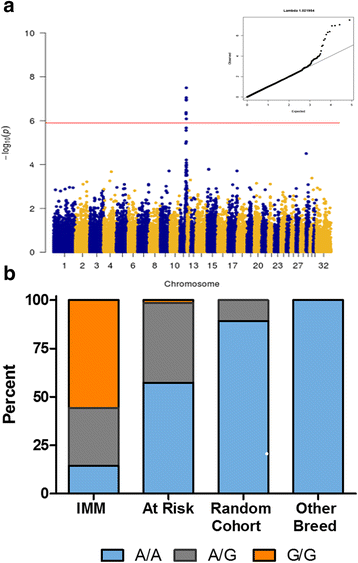
Results: A mixed model analysis identified nine SNPs within a ~ 2.87 Mb region on chr11 that were significantly (Punadjusted < 1.4 × 10- 6) associated with the IMM phenotype. Associated haplotypes within this region encompassed 38 annotated genes, including four myosin genes (MYH1, MYH2, MYH3, and MYH13). Whole genome sequencing of four IMM and four unaffected QHs identified a single segregating nonsynonymous E321G mutation in MYH1 encoding myosin heavy chain 2X. Genotyping of additional 35 IMM and 22 unaffected QHs confirmed an association (P = 2.9 × 10- 5), and the putative mutation was absent in 175 horses from 21 non-QH breeds. Lymphocytic infiltrates occurred in type 2X myofibers and the proportion of 2X fibers was decreased in the presence of inflammation. Protein modeling and contact/stability analysis identified 14 residues affected by the mutation which significantly decreased stability.
Conclusions: We conclude that a mutation in MYH1 is highly associated with susceptibility to the IMM phenotype in QH-related breeds. This is the first report of a mutation in MYH1 and the first link between a skeletal muscle myosin mutation and autoimmune disease.
Keywords: Equine; Genome-wide association; Immunology; Myopathy; Myosin heavy chain 1.
Conflict of interest statement
Ethics approval
All blood samples were collected with approval from the Animal Care and Use Committee at the University of Minnesota and University of California, Davis.
Consent for publication
All data is publically available, and consent for use of all data is available.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Prevalence of the E321G MYH1 variant for immune-mediated myositis and nonexertional rhabdomyolysis in performance subgroups of American Quarter Horses.J Vet Intern Med. 2019 Mar;33(2):897-901. doi: 10.1111/jvim.15393. Epub 2019 Jan 8. J Vet Intern Med. 2019. PMID: 30623495 Free PMC article.
-
An E321G MYH1 mutation is strongly associated with nonexertional rhabdomyolysis in Quarter Horses.J Vet Intern Med. 2018 Sep;32(5):1718-1725. doi: 10.1111/jvim.15299. Epub 2018 Aug 5. J Vet Intern Med. 2018. PMID: 30079499 Free PMC article.
-
Prevalence of the E321G MYH1 variant in Brazilian Quarter Horses.Equine Vet J. 2022 Sep;54(5):952-957. doi: 10.1111/evj.13521. Epub 2021 Oct 28. Equine Vet J. 2022. PMID: 34606642
-
Myosin Heavy Chain Myopathy and Immune-Mediated Muscle Disorders.Vet Clin North Am Equine Pract. 2025 Apr;41(1):61-75. doi: 10.1016/j.cveq.2024.10.005. Epub 2025 Jan 28. Vet Clin North Am Equine Pract. 2025. PMID: 39880733 Review.
-
Immune-Mediated Muscle Diseases of the Horse.Vet Pathol. 2018 Jan;55(1):68-75. doi: 10.1177/0300985816688755. Epub 2017 Jan 27. Vet Pathol. 2018. PMID: 28129093 Review.
Cited by
-
Commercial genetic testing for type 2 polysaccharide storage myopathy and myofibrillar myopathy does not correspond to a histopathological diagnosis.Equine Vet J. 2021 Jul;53(4):690-700. doi: 10.1111/evj.13345. Epub 2020 Oct 29. Equine Vet J. 2021. PMID: 32896939 Free PMC article.
-
Prevalence of the E321G MYH1 variant for immune-mediated myositis and nonexertional rhabdomyolysis in performance subgroups of American Quarter Horses.J Vet Intern Med. 2019 Mar;33(2):897-901. doi: 10.1111/jvim.15393. Epub 2019 Jan 8. J Vet Intern Med. 2019. PMID: 30623495 Free PMC article.
-
Prevalence of clinical signs and factors impacting expression of myosin heavy chain myopathy in Quarter Horse-related breeds with the MYH1E321G mutation.J Vet Intern Med. 2022 May;36(3):1152-1159. doi: 10.1111/jvim.16417. Epub 2022 Apr 14. J Vet Intern Med. 2022. PMID: 35426178 Free PMC article.
-
An E321G MYH1 mutation is strongly associated with nonexertional rhabdomyolysis in Quarter Horses.J Vet Intern Med. 2018 Sep;32(5):1718-1725. doi: 10.1111/jvim.15299. Epub 2018 Aug 5. J Vet Intern Med. 2018. PMID: 30079499 Free PMC article.
-
A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing.DNA Res. 2018 Aug 1;25(4):421-437. doi: 10.1093/dnares/dsy014. DNA Res. 2018. PMID: 29850846 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

