Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection
- PMID: 29511063
- PMCID: PMC5881474
- DOI: 10.1084/jem.20180118
Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection
Abstract
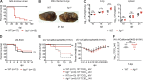
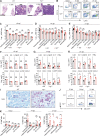
Immune-Responsive Gene 1 (Irg1) is a mitochondrial enzyme that produces itaconate under inflammatory conditions, principally in cells of myeloid lineage. Cell culture studies suggest that itaconate regulates inflammation through its inhibitory effects on cytokine and reactive oxygen species production. To evaluate the functions of Irg1 in vivo, we challenged wild-type (WT) and Irg1-/- mice with Mycobacterium tuberculosis (Mtb) and monitored disease progression. Irg1-/-, but not WT, mice succumbed rapidly to Mtb, and mortality was associated with increased infection, inflammation, and pathology. Infection of LysM-Cre Irg1fl/fl, Mrp8-Cre Irg1fl/fl, and CD11c-Cre Irg1fl/fl conditional knockout mice along with neutrophil depletion experiments revealed a role for Irg1 in LysM+ myeloid cells in preventing neutrophil-mediated immunopathology and disease. RNA sequencing analyses suggest that Irg1 and its production of itaconate temper Mtb-induced inflammatory responses in myeloid cells at the transcriptional level. Thus, an Irg1 regulatory axis modulates inflammation to curtail Mtb-induced lung disease.
© 2018 Nair et al.
Figures




References
-
- Computer Technologies Laboratory 2016. quant3p. A set of scripts to 3′ RNA-seq quantification. Available at: https://github.com/ctlab/quant3p (Accessed February 27, 2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

