Chikungunya Virus Strains Show Lineage-Specific Variations in Virulence and Cross-Protective Ability in Murine and Nonhuman Primate Models
- PMID: 29511072
- PMCID: PMC5844994
- DOI: 10.1128/mBio.02449-17
Chikungunya Virus Strains Show Lineage-Specific Variations in Virulence and Cross-Protective Ability in Murine and Nonhuman Primate Models
Abstract
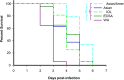
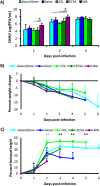
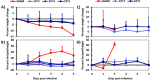
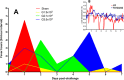
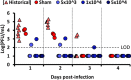
Chikungunya virus (CHIKV) is a reemerging arbovirus capable of causing explosive outbreaks of febrile illness, polyarthritis, and polyarthralgia, inflicting severe morbidity on affected populations. CHIKV can be genetically classified into 3 major lineages: West African (WA); East, Central, and South African (ECSA); Indian Ocean (IOL); and Asian. Additionally, the Indian Ocean (IOL) sublineage emerged within the ECSA clade and the Asian/American sublineage emerged within the Asian clade. While differences in epidemiological and pathological characteristics among outbreaks involving different CHIKV lineages and sublineages have been suggested, few targeted investigations comparing lineage virulence levels have been reported. We compared the virulence levels of CHIKV isolates representing all major lineages and sublineages in the type I interferon receptor-knockout A129 mouse model and found lineage-specific differences in virulence. We also evaluated the cross-protective efficacy of the IOL-derived, live-attenuated vaccine strain CHIKV/IRESv1 against the Asian/American CHIKV isolate YO123223 in both murine and nonhuman primate models, as well as the WA strain SH2830 in a murine model. The CHIKV/IRES vaccine provided protection both in mice and in nonhuman primate cohorts against Caribbean strain challenge and protected mice against WA challenge. Taken together, our data suggest that Asian/American CHIKV strains are less virulent than those in the Asian, ECSA, and WA lineages and that despite differences in virulence, IOL-based vaccine strains offer robust cross-protection against strains from other lineages. Further research is needed to elucidate the genetic basis for variation in CHIKV virulence in the A129 mouse model and to corroborate this variation with human pathogenicity.IMPORTANCE Chikungunya virus (CHIKV) is a reemerging human pathogen capable of causing debilitating and disfiguring polyarthritis, which can last for months to years after initial fever has resolved. There are four major genetic lineages of CHIKV, as well as two recently emerged sublineages, none of which have been evaluated for differences in virulence. Moreover, the ability of chikungunya vaccines to cross-protect against heterologous CHIKV lineages has not been explored. Therefore, we sought to compare the virulence levels among CHIKV lineages, as well as to evaluate the cross-protective efficacy of the CHIKV/IRESv1 vaccine candidate, in two different models of CHIKV infection. Our results suggest that, although significant differences in virulence were observed among CHIKV lineages, the CHIKV/IRESv1 vaccine elicits cross-lineage protective immunity. These findings provide valuable information for predicting the severity of CHIKV-associated morbidity in future outbreaks, as well as vaccine development considerations.
Keywords: alphavirus; chikungunya; vaccine.
Copyright © 2018 Langsjoen et al.
Figures
References
-
- Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, Nasar F, Schuh AJ, Holmes EC, Higgs S, Maharaj PD, Brault AC, Weaver SC. 2010. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol 84:6497–6504. doi: 10.1128/JVI.01603-09. - DOI - PMC - PubMed
-
- Sahadeo N, Mohammed H, Allicock OM, Auguste AJ, Widen SG, Badal K, Pulchan K, Foster JE, Weaver SC, Carrington CV. 2015. Molecular characterisation of Chikungunya virus infections in Trinidad and comparison of clinical and laboratory features with dengue and other acute febrile cases. PLoS Negl Trop Dis 9:e0004199. doi: 10.1371/journal.pntd.0004199. - DOI - PMC - PubMed
-
- Kautz TF, Díaz-González EE, Erasmus JH, Malo-García IR, Langsjoen RM, Patterson EI, Auguste DI, Forrester NL, Sanchez-Casas RM, Hernández-Ávila M, Alpuche-Aranda CM, Weaver SC, Fernández-Salas I. 2015. Chikungunya virus as cause of febrile illness outbreak, Chiapas, Mexico, 2014. Emerg Infect Dis 21:2070–2073. doi: 10.3201/eid2111.150546. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
