Rapid Growth of Uropathogenic Escherichia coli during Human Urinary Tract Infection
- PMID: 29511075
- PMCID: PMC5844997
- DOI: 10.1128/mBio.00186-18
Rapid Growth of Uropathogenic Escherichia coli during Human Urinary Tract Infection
Abstract
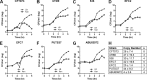
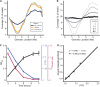
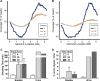
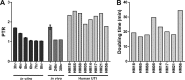
Uropathogenic Escherichia coli (UPEC) strains cause most uncomplicated urinary tract infections (UTIs). These strains are a subgroup of extraintestinal pathogenic E. coli (ExPEC) strains that infect extraintestinal sites, including urinary tract, meninges, bloodstream, lungs, and surgical sites. Here, we hypothesize that UPEC isolates adapt to and grow more rapidly within the urinary tract than other E. coli isolates and survive in that niche. To date, there has not been a reliable method available to measure their growth rate in vivo Here we used two methods: segregation of nonreplicating plasmid pGTR902, and peak-to-trough ratio (PTR), a sequencing-based method that enumerates bacterial chromosomal replication forks present during cell division. In the murine model of UTI, UPEC strain growth was robust in vivo, matching or exceeding in vitro growth rates and only slowing after reaching high CFU counts at 24 and 30 h postinoculation (hpi). In contrast, asymptomatic bacteriuria (ABU) strains tended to maintain high growth rates in vivo at 6, 24, and 30 hpi, and population densities did not increase, suggesting that host responses or elimination limited population growth. Fecal strains displayed moderate growth rates at 6 hpi but did not survive to later times. By PTR, E. coli in urine of human patients with UTIs displayed extraordinarily rapid growth during active infection, with a mean doubling time of 22.4 min. Thus, in addition to traditional virulence determinants, including adhesins, toxins, iron acquisition, and motility, very high growth rates in vivo and resistance to the innate immune response appear to be critical phenotypes of UPEC strains.IMPORTANCE Uropathogenic Escherichia coli (UPEC) strains cause most urinary tract infections in otherwise healthy women. While we understand numerous virulence factors are utilized by E. coli to colonize and persist within the urinary tract, these properties are inconsequential unless bacteria can divide rapidly and survive the host immune response. To determine the contribution of growth rate to successful colonization and persistence, we employed two methods: one involving the segregation of a nonreplicating plasmid in bacteria as they divide and the peak-to-trough ratio, a sequencing-based method that enumerates chromosomal replication forks present during cell division. We found that UPEC strains divide extraordinarily rapidly during human UTIs. These techniques will be broadly applicable to measure in vivo growth rates of other bacterial pathogens during host colonization.
Keywords: ABU; ExPEC; PTR; UPEC; UTI; in vivo growth; plasmid segregation.
Copyright © 2018 Forsyth et al.
Figures

References
-
- Warren JW. 1996. Clinical presentations and epidemiology of urinary tract infections, p 3–27. In Mobley HLT, Warren JW (ed), Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, DC.
-
- Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 169:1–38. - PubMed
-
- Ipe DS, Sundac L, Benjamin WH Jr, Moore KH, Ulett GC. 2013. Asymptomatic bacteriuria: prevalence rates of causal microorganisms, etiology of infection in different patient populations, and recent advances in molecular detection. FEMS Microbiol Lett 346:1–10. doi: 10.1111/1574-6968.12204. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
