Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease
- PMID: 29511076
- PMCID: PMC5844999
- DOI: 10.1128/mBio.00221-18
Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease
Abstract
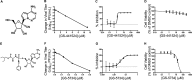
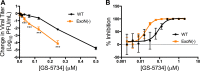
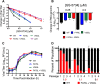
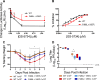
Emerging coronaviruses (CoVs) cause severe disease in humans, but no approved therapeutics are available. The CoV nsp14 exoribonuclease (ExoN) has complicated development of antiviral nucleosides due to its proofreading activity. We recently reported that the nucleoside analogue GS-5734 (remdesivir) potently inhibits human and zoonotic CoVs in vitro and in a severe acute respiratory syndrome coronavirus (SARS-CoV) mouse model. However, studies with GS-5734 have not reported resistance associated with GS-5734, nor do we understand the action of GS-5734 in wild-type (WT) proofreading CoVs. Here, we show that GS-5734 inhibits murine hepatitis virus (MHV) with similar 50% effective concentration values (EC50) as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). Passage of WT MHV in the presence of the GS-5734 parent nucleoside selected two mutations in the nsp12 polymerase at residues conserved across all CoVs that conferred up to 5.6-fold resistance to GS-5734, as determined by EC50 The resistant viruses were unable to compete with WT in direct coinfection passage in the absence of GS-5734. Introduction of the MHV resistance mutations into SARS-CoV resulted in the same in vitro resistance phenotype and attenuated SARS-CoV pathogenesis in a mouse model. Finally, we demonstrate that an MHV mutant lacking ExoN proofreading was significantly more sensitive to GS-5734. Combined, the results indicate that GS-5734 interferes with the nsp12 polymerase even in the setting of intact ExoN proofreading activity and that resistance can be overcome with increased, nontoxic concentrations of GS-5734, further supporting the development of GS-5734 as a broad-spectrum therapeutic to protect against contemporary and emerging CoVs.IMPORTANCE Coronaviruses (CoVs) cause severe human infections, but there are no approved antivirals to treat these infections. Development of nucleoside-based therapeutics for CoV infections has been hampered by the presence of a proofreading exoribonuclease. Here, we expand the known efficacy of the nucleotide prodrug remdesivir (GS-5734) to include a group β-2a CoV. Further, GS-5734 potently inhibits CoVs with intact proofreading. Following selection with the GS-5734 parent nucleoside, 2 amino acid substitutions in the nsp12 polymerase at residues that are identical across CoVs provide low-level resistance to GS-5734. The resistance mutations decrease viral fitness of MHV in vitro and attenuate pathogenesis in a SARS-CoV animal model of infection. Together, these studies define the target of GS-5734 activity and demonstrate that resistance is difficult to select, only partial, and impairs fitness and virulence of MHV and SARS-CoV, supporting further development of GS-5734 as a potential effective pan-CoV antiviral.
Keywords: RNA polymerases; SARS-CoV; antiviral agents; antiviral resistance; coronavirus; nucleoside analogs; pandemic.
Copyright © 2018 Agostini et al.
Figures



References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A, Humphrey CD, Shieh W, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

