Mechanism of Darunavir (DRV)'s High Genetic Barrier to HIV-1 Resistance: A Key V32I Substitution in Protease Rarely Occurs, but Once It Occurs, It Predisposes HIV-1 To Develop DRV Resistance
- PMID: 29511083
- PMCID: PMC5844992
- DOI: 10.1128/mBio.02425-17
Mechanism of Darunavir (DRV)'s High Genetic Barrier to HIV-1 Resistance: A Key V32I Substitution in Protease Rarely Occurs, but Once It Occurs, It Predisposes HIV-1 To Develop DRV Resistance
Abstract
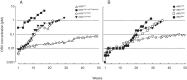
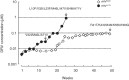
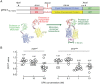
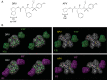
Darunavir (DRV) has bimodal activity against HIV-1 protease, enzymatic inhibition and protease dimerization inhibition, and has an extremely high genetic barrier against development of drug resistance. We previously generated a highly DRV-resistant HIV-1 variant (HIVDRVRP51). We also reported that four amino acid substitutions (V32I, L33F, I54M, and I84V) identified in the protease of HIVDRVRP51 are largely responsible for its high-level resistance to DRV. Here, we attempted to elucidate the role of each of the four amino acid substitutions in the development of DRV resistance. We found that V32I is a key substitution, which rarely occurs, but once it occurs, it predisposes HIV-1 to develop high-level DRV resistance. When two infectious recombinant HIV-1 clones carrying I54M and I84V (rHIVI54M and rHIVI84V, respectively) were selected in the presence of DRV, V32I emerged, and the virus rapidly developed high-level DRV resistance. rHIVV32I also developed high-level DRV resistance. However, wild-type HIVNL4-3 (rHIVWT) failed to acquire V32I and did not develop DRV resistance. Compared to rHIVWT, rHIVV32I was highly susceptible to DRV and had significantly reduced fitness, explaining why V32I did not emerge upon selection of rHIVWT with DRV. When the only substitution is at residue 32, structural analysis revealed much stronger van der Waals interactions between DRV and I-32 than between DRV and V-32. These results suggest that V32I is a critical amino acid substitution in multiple pathways toward HIV-1's DRV resistance development and elucidate, at least in part, a mechanism of DRV's high genetic barrier to development of drug resistance. The results also show that attention should be paid to the initiation or continuation of DRV-containing regimens in people with HIV-1 containing the V32I substitution.IMPORTANCE Darunavir (DRV) is the only protease inhibitor (PI) recommended as a first-line therapeutic and represents the most widely used PI for treating HIV-1-infected individuals. DRV possesses a high genetic barrier to development of HIV-1's drug resistance. However, the mechanism(s) of the DRV's high genetic barrier remains unclear. Here, we show that the preexistence of certain single amino acid substitutions such as V32I, I54M, A71V, and I84V in HIV-1 protease facilitates the development of high-level DRV resistance. Interestingly, all in vitro-selected highly DRV-resistant HIV-1 variants acquired V32I but never emerged in wild-type HIV (HIVWT), and V32I itself rendered HIV-1 more sensitive to DRV and reduced viral fitness compared to HIVWT, strongly suggesting that the emergence of V32I plays a critical role in the development of HIV-1's resistance to DRV. Our results would be of benefit in the treatment of HIV-1-infected patients receiving DRV-containing regimens.
Keywords: HIV-1; V32I; darunavir; drug resistance; dual mechanism; genetic barrier; protease inhibitors.
Figures



References
-
- Samji H, Cescon A, Hogg RS, Modur SP, Althoff KN, Buchacz K, Burchell AN, Cohen M, Gebo KA, Gill MJ, Justice A, Kirk G, Klein MB, Korthuis PT, Martin J, Napravnik S, Rourke SB, Sterling TR, Silverberg MJ, Deeks S, Jacobson LP, Bosch RJ, Kitahata MM, Goedert JJ, Moore R, Gange SJ, North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) of IeDEA . 2013. Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS One 8:e81355. doi:10.1371/journal.pone.0081355. - DOI - PMC - PubMed
-
- Marcus JL, Chao CR, Leyden WA, Xu L, Quesenberry CP Jr, Klein DB, Towner WJ, Horberg MA, Silverberg MJ. 2016. Narrowing the gap in life expectancy between HIV-infected and HIV-uninfected individuals with access to care. J Acquir Immune Defic Syndr 73:39–46. doi:10.1097/QAI.0000000000001014. - DOI - PMC - PubMed
-
- DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents 2017. Guidelines for the use of antiretroviral agents in adults and adolescents living with HIV. DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents, Office of AIDS Research Advisory Council, US Department of Health and Human Services, Washington, DC.
-
- Koh Y, Matsumi S, Das D, Amano M, Davis DA, Li J, Leschenko S, Baldridge A, Shioda T, Yarchoan R, Ghosh AK, Mitsuya H. 2007. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J Biol Chem 282:28709–28720. doi:10.1074/jbc.M703938200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
