Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: comparison of Laemmli and TRIzol© protein-extraction methods
- PMID: 29511462
- PMCID: PMC5827780
- DOI: 10.1080/20013078.2018.1438727
Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: comparison of Laemmli and TRIzol© protein-extraction methods
Abstract
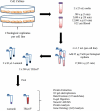
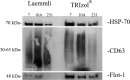
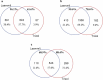
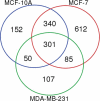
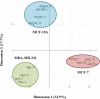
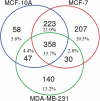
Sample amount is often a limiting factor for multi-parametric analyses that encompass at least three areas of '-omics' research: genomics, transcriptomics and proteomics. Limited sample amounts are also an important consideration when these multi-parametric analyses are performed on extracellular vesicles (EVs), as the amount of EVs (and EV cargo) that can be isolated is often very low. It is well understood that a monophasic solution of phenol and guanidine isothiocyanate (i.e. TRIzol©) can simultaneously isolate DNA, RNA and proteins from biological samples; however, it is most commonly used for the extraction of RNA. Validation of this reagent for the isolation of multiple classes of biological molecules from EVs would provide a widely applicable method for performing multi-parametric analyses of EV material. In this report, we describe a comparison of proteins identified from EVs processed with either TRIzol© or the conventional Laemmli buffer protein-extraction reagents. EVs were isolated from 3 mL of cell-culture supernatant derived from MCF-10A, MCF-7 and MDA-MB-231 cells using the Vn96 EV capture technology. For the TRIzol© extraction protocol, proteins were precipitated with acetone from the organic phase and then re-solubilized in a mixture of 8M urea, 0.2% SDS and 1 M Tris-HCl pH 6.8, followed by dilution in 5× loading buffer prior to fractionation with 1D SDS-PAGE. NanoLC-MS/MS of the trypsin-digested proteins was used to generate proteomic profiles from EV protein samples extracted with each method. Of the identified proteins, 57.7%, 69.2% and 57.0% were common to both extraction methods for EVs from MCF-10A, MCF-7 and MDA-MB-231, respectively. Our results suggest that TRIzol© extraction of proteins from EVs has significant equivalence to the traditional Laemmli method. The advantage of using TRIzol
Keywords: MCF-10A; MCF-7; MDA-MB-231; Vn96; cell-conditioned media; exosomes; extracellular vesicle.
© reagent is the ability to accumulate multi-parametric data (e.g., RNA and protein profiles) on the same limited EV sample while minimizing sample preparation and processing time.
Conflict of interest statement
The Vn96 EV extraction reagent and protocol were developed by the Atlantic Cancer Research Institute and commercialized in partnership with New England Peptide (Gardner, MA).
Figures
Similar articles
-
A multiparametric extraction method for Vn96-isolated plasma extracellular vesicles and cell-free DNA that enables multi-omic profiling.Sci Rep. 2021 Apr 13;11(1):8085. doi: 10.1038/s41598-021-87526-y. Sci Rep. 2021. PMID: 33850235 Free PMC article.
-
Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method.J Extracell Vesicles. 2017 Apr 28;6(1):1313091. doi: 10.1080/20013078.2017.1313091. eCollection 2017. J Extracell Vesicles. 2017. PMID: 28717416 Free PMC article.
-
Peptide-Affinity Isolation of Extracellular Vesicles and Cell-Free DNA From Human Plasma.Methods Mol Biol. 2022;2508:341-352. doi: 10.1007/978-1-0716-2376-3_22. Methods Mol Biol. 2022. PMID: 35737249
-
Chromatography and its hyphenation to mass spectrometry for extracellular vesicle analysis.J Chromatogr A. 2016 Mar 25;1439:26-41. doi: 10.1016/j.chroma.2016.01.017. Epub 2016 Jan 11. J Chromatogr A. 2016. PMID: 26830636 Review.
-
The Role of Extracellular Vesicles in Diseases of the Ear, Nose, and Throat.Med Sci (Basel). 2022 Dec 28;11(1):6. doi: 10.3390/medsci11010006. Med Sci (Basel). 2022. PMID: 36649043 Free PMC article. Review.
Cited by
-
Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice.Plant Biotechnol J. 2021 Nov;19(11):2304-2318. doi: 10.1111/pbi.13661. Epub 2021 Aug 24. Plant Biotechnol J. 2021. PMID: 34245650 Free PMC article.
-
Progress in exosome associated tumor markers and their detection methods.Mol Biomed. 2020 Aug 14;1(1):3. doi: 10.1186/s43556-020-00002-3. Mol Biomed. 2020. PMID: 35006428 Free PMC article. Review.
-
Flow Cytometry Analysis of Circulating Extracellular Vesicle Subtypes from Fresh Peripheral Blood Samples.Int J Mol Sci. 2020 Dec 23;22(1):48. doi: 10.3390/ijms22010048. Int J Mol Sci. 2020. PMID: 33374539 Free PMC article.
-
A multiparametric extraction method for Vn96-isolated plasma extracellular vesicles and cell-free DNA that enables multi-omic profiling.Sci Rep. 2021 Apr 13;11(1):8085. doi: 10.1038/s41598-021-87526-y. Sci Rep. 2021. PMID: 33850235 Free PMC article.
-
The Potential of Aptamer-Mediated Liquid Biopsy for Early Detection of Cancer.Int J Mol Sci. 2021 May 25;22(11):5601. doi: 10.3390/ijms22115601. Int J Mol Sci. 2021. PMID: 34070509 Free PMC article. Review.
References
-
- Palazzolo G, Albanese NN, Cara GD, et al. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012;32(3):847–860. - PubMed
-
- Revenfeld ALS, Baek R, Nielsen MH, et al. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Therapeut. 2014;36(6):830–846. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

