Adenovirus-Mediated Delivery of Decoy Hyper Binding Sites Targeting Oncogenic HMGA1 Reduces Pancreatic and Liver Cancer Cell Viability
- PMID: 29511732
- PMCID: PMC5832671
- DOI: 10.1016/j.omto.2018.01.002
Adenovirus-Mediated Delivery of Decoy Hyper Binding Sites Targeting Oncogenic HMGA1 Reduces Pancreatic and Liver Cancer Cell Viability
Abstract
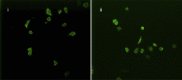
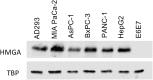
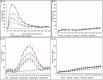
High mobility group AT-hook 1 (HMGA1) protein is an oncogenic architectural transcription factor that plays an essential role in early development, but it is also implicated in many human cancers. Elevated levels of HMGA1 in cancer cells cause misregulation of gene expression and are associated with increased cancer cell proliferation and increased chemotherapy resistance. We have devised a strategy of using engineered viruses to deliver decoy hyper binding sites for HMGA1 to the nucleus of cancer cells with the goal of sequestering excess HMGA1 at the decoy hyper binding sites due to binding competition. Sequestration of excess HMGA1 at the decoy binding sites is intended to reduce HMGA1 binding at the naturally occurring genomic HMGA1 binding sites, which should result in normalized gene expression and restored sensitivity to chemotherapy. As proof of principle, we engineered the replication defective adenovirus serotype 5 genome to contain hyper binding sites for HMGA1 composed of six copies of an individual HMGA1 binding site, referred to as HMGA-6. A 70%-80% reduction in cell viability and increased sensitivity to gemcitabine was observed in five different pancreatic and liver cancer cell lines 72 hr after infection with replication defective engineered adenovirus serotype 5 virus containing the HMGA-6 decoy hyper binding sites. The decoy hyper binding site strategy should be general for targeting overexpression of any double-stranded DNA-binding oncogenic transcription factor responsible for cancer cell proliferation.
Keywords: HMGA1; adenovirus; cancer therapy; chemotherapy resistance; decoy binding site; high mobility group A protein; liver cancer; neoadjuvant therapy; oncogenic transcription factor; pancreatic cancer.
Figures





Similar articles
-
A mouse model study of toxicity and biodistribution of a replication defective adenovirus serotype 5 virus with its genome engineered to contain a decoy hyper binding site to sequester and suppress oncogenic HMGA1 as a new cancer treatment therapy.PLoS One. 2018 Feb 20;13(2):e0192882. doi: 10.1371/journal.pone.0192882. eCollection 2018. PLoS One. 2018. PMID: 29462157 Free PMC article.
-
HMGA1 expression levels are elevated in pancreatic intraepithelial neoplasia cells in the Ptf1a-Cre; LSL-KrasG12D transgenic mouse model of pancreatic cancer.Br J Cancer. 2017 Aug 22;117(5):639-647. doi: 10.1038/bjc.2017.216. Epub 2017 Jul 11. Br J Cancer. 2017. PMID: 28697176 Free PMC article.
-
Characterization of the Stoichiometry of HMGA1/DNA Complexes.Open Biochem J. 2013 Sep 4;7:73-81. doi: 10.2174/1874091X01307010073. eCollection 2013. Open Biochem J. 2013. PMID: 24062859 Free PMC article.
-
RNA-Mediated Regulation of HMGA1 Function.Biomolecules. 2015;5(2):943-57. doi: 10.3390/biom5020943. Biomolecules. 2015. PMID: 26117853 Free PMC article. Review.
-
HMGA1-pseudogene overexpression contributes to cancer progression.Cell Cycle. 2014;13(23):3636-9. doi: 10.4161/15384101.2014.974440. Cell Cycle. 2014. PMID: 25483074 Free PMC article. Review.
Cited by
-
Harnessing the Therapeutic Potential of Decoys in Non-Atherosclerotic Cardiovascular Diseases: State of the Art.J Cardiovasc Dev Dis. 2021 Aug 27;8(9):103. doi: 10.3390/jcdd8090103. J Cardiovasc Dev Dis. 2021. PMID: 34564121 Free PMC article. Review.
-
The Clinical Potential of Oligonucleotide Therapeutics against Pancreatic Cancer.Int J Mol Sci. 2019 Jul 6;20(13):3331. doi: 10.3390/ijms20133331. Int J Mol Sci. 2019. PMID: 31284594 Free PMC article. Review.
-
A mouse model study of toxicity and biodistribution of a replication defective adenovirus serotype 5 virus with its genome engineered to contain a decoy hyper binding site to sequester and suppress oncogenic HMGA1 as a new cancer treatment therapy.PLoS One. 2018 Feb 20;13(2):e0192882. doi: 10.1371/journal.pone.0192882. eCollection 2018. PLoS One. 2018. PMID: 29462157 Free PMC article.
-
Zinc Oxide Nanocrystals and High-Energy Shock Waves: A New Synergy for the Treatment of Cancer Cells.Front Bioeng Biotechnol. 2020 Jun 5;8:577. doi: 10.3389/fbioe.2020.00577. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32582682 Free PMC article.
-
How Early Can Pancreatic Tumors Be Detected Using NMR-Based Urine Metabolic Profiling? Identification of Early-Stage Biomarkers of Tumor Initiation and Progression in an Orthotopic Xenograft Mouse Model of Pancreatic Cancer.Metabolites. 2025 Feb 20;15(3):142. doi: 10.3390/metabo15030142. Metabolites. 2025. PMID: 40137107 Free PMC article.
References
-
- Reeves R., Nissen M.S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990;265:8573–8582. - PubMed
-
- Huth J.R., Bewley C.A., Nissen M.S., Evans J.N., Reeves R., Gronenborn A.M., Clore G.M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

