Identification of key genes and long non‑coding RNAs in celecoxib‑treated lung squamous cell carcinoma cell line by RNA‑sequencing
- PMID: 29512696
- PMCID: PMC5928627
- DOI: 10.3892/mmr.2018.8656
Identification of key genes and long non‑coding RNAs in celecoxib‑treated lung squamous cell carcinoma cell line by RNA‑sequencing
Abstract
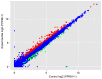
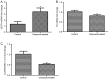
Celecoxib is an inhibitor of cyclooxygenase-2, a gene that is often aberrantly expressed in the lung squamous cell carcinoma (LSQCC). The present study aims to provide novel insight into chemoprevention by celecoxib treatment. The human LSQCC cell line SK‑MES‑1 was treated with or without celecoxib and RNA‑sequencing (RNA‑seq) was performed on the Illumina HiSeq 2000 platform. Expression levels of genes or long non‑coding RNAs (lncRNAs) were calculated by Cufflinks software. Subsequently, differentially expressed genes (DEGs) and differentially expressed lncRNAs (DE‑LNRs) between the two groups were selected using the limma package and LNCipedia 3.0, respectively; followed by co‑expression analysis based on their expression correlation coefficient (CC). Enrichment analysis for the DEGs and co‑expressed DE‑LNRs were performed. Protein‑protein interaction (PPI) network analysis for DEGs was performed using STRING database. A set of 317 DEGs and 25 DE‑LNRs were identified between celecoxib‑treated and non‑treated cell lines. A total of 12 pathways were enriched by the DEGs, including 'protein processing in endoplasmic reticulum' for activating transcription factor 4 (ATF4), 'mammalian target of rapamycin (mTOR) signaling pathway' for vascular endothelial growth factor A (VEGFA) and 'ECM‑receptor interaction' for fibronectin 1 (FN1). Genes such as VEGFA, ATF4 and FN1 were highlighted in the PPI network. VEGFA was linked with lnc‑AP000769.1‑2:10 (CC= ‑0.99227), whereas ATF4 and FN1 were closely correlated with lnc‑HFE2‑2:1 (CC=0.996159 and ‑0.98714, respectively). lncRNAs were also enriched in pathways such as 'mTOR signaling pathway' for lnc‑HFE2‑2:1. Several important molecules were identified in celecoxib‑treated LSQCC cell lines, such as VEGFA, ATF4, FN1, lnc‑AP000769.1‑2:10 and lnc‑HFE2‑2:1, which may enhance the anti‑cancer effects of celecoxib on LSQCC.
Keywords: lung squamous cell carcinoma; celecoxib; long noncoding RNA; chemosensitivity; RNA sequencing; mammalian target of rapamycin signaling.
Figures

References
-
- Bendale Y, Bendale V, Birari-Gawande P, Kadam A, Gund P. Tumor regression with ayurvedic rasayana therapy in squamous cell carcinoma of lungs. Rasamruta. 2015;7:1–5.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

