ETS1 and SP1 drive DHX15 expression in acute lymphoblastic leukaemia
- PMID: 29512921
- PMCID: PMC5908128
- DOI: 10.1111/jcmm.13525
ETS1 and SP1 drive DHX15 expression in acute lymphoblastic leukaemia
Abstract
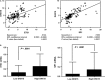
DHX15 plays a role in leukaemogenesis and leukaemia relapse. However, the mechanism underlying the transcriptional regulation of DHX15 in ALL has not been elucidated. Our present study aimed to explore the functional promoter region of DHX15 and to investigate the transcription factors controlling the transcription of this gene. A luciferase assay performed with several truncated constructs identified a 501-bp region as the core promoter region of DHX15. Site-directed mutagenesis, electrophoretic mobility shift and chromatin immunoprecipitation assays showed that ETS1 and SP1 occupied the DHX15 promoter. Furthermore, knockdown of ETS1 and SP1 resulted in suppression of DHX15, whereas the overexpression of these genes led to up-regulation of DHX15. Interestingly, in samples obtained from patients with ALL at diagnosis, both ETS1 and SP1 correlated positively with DHX15 expression. Additionally, differences in methylation of the DHX15 core promoter region were not observed between the patients and controls. In conclusion, we identified the core promoter region of DHX15 and demonstrated that ETS1 and SP1 regulated DHX15 expression in ALL.
Keywords: DHX15; ETS1; SP1; promoter.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures




References
-
- Jabbour EJ, Faderl S, Kantarjian HM. Adult acute lymphoblastic leukemia. Mayo Clin Proc. 2005;80:1517‐1527. - PubMed
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535‐1548. - PubMed
-
- Crazzolara R, Kreczy A, Mann G, et al. High expression of the chemokine receptor CXCR4 predicts extramedullary organ infiltration in childhood acute lymphoblastic leukaemia. Br J Haematol. 2001;115:545‐553. - PubMed
-
- Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75‐87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

