Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte
- PMID: 29513657
- PMCID: PMC5856367
- DOI: 10.1038/nature25964
Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte
Abstract
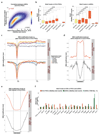
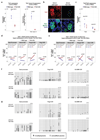
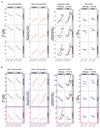
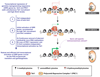
Gametes are highly specialized cells that can give rise to the next generation through their ability to generate a totipotent zygote. In mice, germ cells are first specified in the developing embryo around embryonic day (E) 6.25 as primordial germ cells (PGCs). Following subsequent migration into the developing gonad, PGCs undergo a wave of extensive epigenetic reprogramming around E10.5-E11.5, including genome-wide loss of 5-methylcytosine. The underlying molecular mechanisms of this process have remained unclear, leading to our inability to recapitulate this step of germline development in vitro. Here we show, using an integrative approach, that this complex reprogramming process involves coordinated interplay among promoter sequence characteristics, DNA (de)methylation, the polycomb (PRC1) complex and both DNA demethylation-dependent and -independent functions of TET1 to enable the activation of a critical set of germline reprogramming-responsive genes involved in gamete generation and meiosis. Our results also reveal an unexpected role for TET1 in maintaining but not driving DNA demethylation in gonadal PGCs. Collectively, our work uncovers a fundamental biological role for gonadal germline reprogramming and identifies the epigenetic principles of the PGC-to-gonocyte transition that will help to guide attempts to recapitulate complete gametogenesis in vitro.
Conflict of interest statement
The authors declare no conflict of interest or competing financial interest.
Figures











Comment in
-
Reprogramming: Methylation patterns in primordial germ cells.Nat Rev Mol Cell Biol. 2018 May;19(5):278-279. doi: 10.1038/nrm.2018.20. Epub 2018 Mar 21. Nat Rev Mol Cell Biol. 2018. PMID: 29559735 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

