ERRγ Promotes Angiogenesis, Mitochondrial Biogenesis, and Oxidative Remodeling in PGC1α/β-Deficient Muscle
- PMID: 29514081
- PMCID: PMC5860878
- DOI: 10.1016/j.celrep.2018.02.047
ERRγ Promotes Angiogenesis, Mitochondrial Biogenesis, and Oxidative Remodeling in PGC1α/β-Deficient Muscle
Abstract
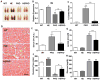
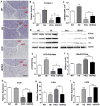
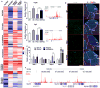
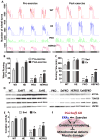
PGC1α is a pleiotropic co-factor that affects angiogenesis, mitochondrial biogenesis, and oxidative muscle remodeling via its association with multiple transcription factors, including the master oxidative nuclear receptor ERRγ. To decipher their epistatic relationship, we explored ERRγ gain of function in muscle-specific PGC1α/β double-knockout (PKO) mice. ERRγ-driven transcriptional reprogramming largely rescues muscle damage and improves muscle function in PKO mice, inducing mitochondrial biogenesis, antioxidant defense, angiogenesis, and a glycolytic-to-oxidative fiber-type transformation independent of PGC1α/β. Furthermore, in combination with voluntary exercise, ERRγ gain of function largely restores mitochondrial energetic deficits in PKO muscle, resulting in a 5-fold increase in running performance. Thus, while PGC1s can interact with multiple transcription factors, these findings implicate ERRs as the major molecular target through which PGC1α/β regulates both innate and adaptive energy metabolism.
Keywords: ERR; PGC1; estrogen related receptor; exercise; fatty acid oxidation; glycolysis; mitochondria; muscle; muscle damage; vasculature.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

