An Essential Role for ECSIT in Mitochondrial Complex I Assembly and Mitophagy in Macrophages
- PMID: 29514094
- PMCID: PMC5909989
- DOI: 10.1016/j.celrep.2018.02.051
An Essential Role for ECSIT in Mitochondrial Complex I Assembly and Mitophagy in Macrophages
Abstract
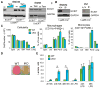
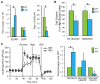
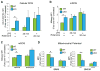
ECSIT is a mitochondrial complex I (CI)-associated protein that has been shown to regulate the production of mitochondrial reactive oxygen species (mROS) following engagement of Toll-like receptors (TLRs). We have generated an Ecsit conditional knockout (CKO) mouse strain to study the in vivo role of ECSIT. ECSIT deletion results in profound alteration of macrophage metabolism, leading to a striking shift to reliance on glycolysis, complete disruption of CI activity, and loss of the CI holoenzyme and multiple subassemblies. An increase in constitutive mROS production in ECSIT-deleted macrophages prevents further TLR-induced mROS production. Surprisingly, ECSIT-deleted cells accumulate damaged mitochondria because of defective mitophagy. ECSIT associates with the mitophagy regulator PINK1 and exhibits Parkin-dependent ubiquitination. However, upon ECSIT deletion, we observed increased mitochondrial Parkin without the expected increase in mitophagy. Taken together, these results demonstrate a key role of ECSIT in CI function, mROS production, and mitophagy-dependent mitochondrial quality control.
Keywords: ROS; complex I; glycolytic switch; mROS; macrophages; mitophagy; oxidative stress.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10–12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

