Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation
- PMID: 29514250
- PMCID: PMC5961081
- DOI: 10.1093/nar/gky160
Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation
Abstract
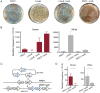
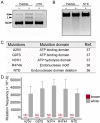
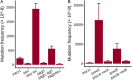
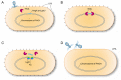
Pseudomonas aeruginosa is an opportunistic pathogen with a relatively large genome, and has been shown to routinely lose genomic fragments during environmental selection. However, the underlying molecular mechanisms that promote chromosomal deletion are still poorly understood. In a recent study, we showed that by deleting a large chromosomal fragment containing two closely situated genes, hmgA and galU, P. aeruginosa was able to form 'brown mutants', bacteriophage (phage) resistant mutants with a brown color phenotype. In this study, we show that the brown mutants occur at a frequency of 227 ± 87 × 10-8 and contain a deletion ranging from ∼200 to ∼620 kb. By screening P. aeruginosa transposon mutants, we identified mutL gene whose mutation constrained the emergence of phage-resistant brown mutants. Moreover, the P. aeruginosa MutL (PaMutL) nicking activity can result in DNA double strand break (DSB), which is then repaired by non-homologous end joining (NHEJ), leading to chromosomal deletions. Thus, we reported a noncanonical function of PaMutL that promotes chromosomal deletions through NHEJ to prevent phage predation.
Figures

Similar articles
-
Fluctuating Bacteriophage-induced galU Deficiency Region is Involved in Trade-off Effects on the Phage and Fluoroquinolone Sensitivity in Pseudomonas aeruginosa.Virus Res. 2021 Dec;306:198596. doi: 10.1016/j.virusres.2021.198596. Epub 2021 Oct 12. Virus Res. 2021. PMID: 34648885
-
Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa.Sci Rep. 2014 Apr 28;4:4738. doi: 10.1038/srep04738. Sci Rep. 2014. PMID: 24770387 Free PMC article.
-
Increased Innate Immune Susceptibility in Hyperpigmented Bacteriophage-Resistant Mutants of Pseudomonas aeruginosa.Antimicrob Agents Chemother. 2022 Aug 16;66(8):e0023922. doi: 10.1128/aac.00239-22. Epub 2022 Jul 6. Antimicrob Agents Chemother. 2022. PMID: 35862755 Free PMC article.
-
Microhomology-mediated end joining: Good, bad and ugly.Mutat Res. 2018 May;809:81-87. doi: 10.1016/j.mrfmmm.2017.07.002. Epub 2017 Jul 16. Mutat Res. 2018. PMID: 28754468 Free PMC article. Review.
-
Regulation of repair pathway choice at two-ended DNA double-strand breaks.Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review.
Cited by
-
Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy.Int J Mol Sci. 2023 Oct 26;24(21):15628. doi: 10.3390/ijms242115628. Int J Mol Sci. 2023. PMID: 37958612 Free PMC article. Review.
-
Combined Bacteriophage and Antibiotic Treatment Prevents Pseudomonas aeruginosa Infection of Wild Type and cftr- Epithelial Cells.Front Microbiol. 2020 Aug 26;11:1947. doi: 10.3389/fmicb.2020.01947. eCollection 2020. Front Microbiol. 2020. PMID: 32983005 Free PMC article.
-
A Combination of Virulent and Non-Productive Phages Synergizes the Immune System against Salmonella Typhimurium Systemic Infection.Int J Mol Sci. 2022 Oct 24;23(21):12830. doi: 10.3390/ijms232112830. Int J Mol Sci. 2022. PMID: 36361621 Free PMC article.
-
Phages ZC01 and ZC03 require type-IV pilus for Pseudomonas aeruginosa infection and have a potential for therapeutic applications.Microbiol Spectr. 2024 Oct 29;12(12):e0152724. doi: 10.1128/spectrum.01527-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39470273 Free PMC article.
-
RNA and Single-Stranded DNA Phages: Unveiling the Promise from the Underexplored World of Viruses.Int J Mol Sci. 2023 Dec 1;24(23):17029. doi: 10.3390/ijms242317029. Int J Mol Sci. 2023. PMID: 38069353 Free PMC article. Review.
References
-
- Moran N.A. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002; 108:583–586. - PubMed
-
- Hocquet D., Petitjean M., Rohmer L., Valot B., Kulasekara H.D., Bedel E., Bertrand X., Plesiat P., Kohler T., Pantel A. et al. . Pyomelanin-producing Pseudomonas aeruginosa selected during chronic infections have a large chromosomal deletion which confers resistance to pyocins. Environ. Microbiol. 2016; 18:3482–3493. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

