In cell mutational interference mapping experiment (in cell MIME) identifies the 5' polyadenylation signal as a dual regulator of HIV-1 genomic RNA production and packaging
- PMID: 29514260
- PMCID: PMC5961354
- DOI: 10.1093/nar/gky152
In cell mutational interference mapping experiment (in cell MIME) identifies the 5' polyadenylation signal as a dual regulator of HIV-1 genomic RNA production and packaging
Abstract
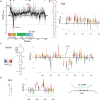
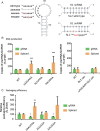
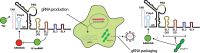
Non-coding RNA regulatory elements are important for viral replication, making them promising targets for therapeutic intervention. However, regulatory RNA is challenging to detect and characterise using classical structure-function assays. Here, we present in cell Mutational Interference Mapping Experiment (in cell MIME) as a way to define RNA regulatory landscapes at single nucleotide resolution under native conditions. In cell MIME is based on (i) random mutation of an RNA target, (ii) expression of mutated RNA in cells, (iii) physical separation of RNA into functional and non-functional populations, and (iv) high-throughput sequencing to identify mutations affecting function. We used in cell MIME to define RNA elements within the 5' region of the HIV-1 genomic RNA (gRNA) that are important for viral replication in cells. We identified three distinct RNA motifs controlling intracellular gRNA production, and two distinct motifs required for gRNA packaging into virions. Our analysis reveals the 73AAUAAA78 polyadenylation motif within the 5' PolyA domain as a dual regulator of gRNA production and gRNA packaging, and demonstrates that a functional polyadenylation signal is required for viral packaging even though it negatively affects gRNA production.
Figures


Similar articles
-
An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation.Retrovirology. 2018 Mar 14;15(1):25. doi: 10.1186/s12977-018-0407-4. Retrovirology. 2018. PMID: 29540207 Free PMC article.
-
HIV-1 integrase binding to genomic RNA 5'-UTR induces local structural changes in vitro and in virio.Retrovirology. 2021 Nov 22;18(1):37. doi: 10.1186/s12977-021-00582-0. Retrovirology. 2021. PMID: 34809662 Free PMC article.
-
A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation.Cell Host Microbe. 2013 Feb 13;13(2):181-92. doi: 10.1016/j.chom.2013.01.007. Cell Host Microbe. 2013. PMID: 23414758 Free PMC article.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
-
NMR Studies of the Structure and Function of the HIV-1 5'-Leader.Viruses. 2016 Dec 21;8(12):338. doi: 10.3390/v8120338. Viruses. 2016. PMID: 28009832 Free PMC article. Review.
Cited by
-
RNA Structures and Their Role in Selective Genome Packaging.Viruses. 2021 Sep 8;13(9):1788. doi: 10.3390/v13091788. Viruses. 2021. PMID: 34578369 Free PMC article. Review.
-
ITN-VIROINF: Understanding (Harmful) Virus-Host Interactions by Linking Virology and Bioinformatics.Viruses. 2021 Apr 27;13(5):766. doi: 10.3390/v13050766. Viruses. 2021. PMID: 33925452 Free PMC article.
-
Expression, purification, and characterization of biologically active full-length Mason-Pfizer monkey virus (MPMV) Pr78Gag.Sci Rep. 2018 Aug 7;8(1):11793. doi: 10.1038/s41598-018-30142-0. Sci Rep. 2018. PMID: 30087395 Free PMC article.
-
Expression, purification, and functional characterization of soluble recombinant full-length simian immunodeficiency virus (SIV) Pr55Gag.Heliyon. 2023 Jan 10;9(1):e12892. doi: 10.1016/j.heliyon.2023.e12892. eCollection 2023 Jan. Heliyon. 2023. PMID: 36685375 Free PMC article.
-
RanDeL-Seq: a High-Throughput Method to Map Viral cis- and trans-Acting Elements.mBio. 2021 Jan 19;12(1):e01724-20. doi: 10.1128/mBio.01724-20. mBio. 2021. PMID: 33468683 Free PMC article.
References
-
- Paillart J.C., Dettenhofer M., Yu X.F., Ehresmann C., Ehresmann B., Marquet R.. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004; 279:48397–48403. - PubMed
-
- Abd El-Wahab E.W., Smyth R.P., Mailler E., Bernacchi S., Vivet-Boudou V., Hijnen M., Jossinet F., Mak J., Paillart J.-C., Marquet R.. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 2014; 5:4304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

